Probe CUST_43302_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43302_PI426222305 | JHI_St_60k_v1 | DMT400064456 | TGACACTCTTGTGTTCAAGTACATGAAGGGATCAAACTCAGTATTAGTGGTAAACAAGGA |
All Microarray Probes Designed to Gene DMG400025042
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43323_PI426222305 | JHI_St_60k_v1 | DMT400064455 | TACCGGCTCAAATAACTCGGCAGTAAAAGCATTTACTACAACTTCAGCTGTTTTTGTGTC |
| CUST_43302_PI426222305 | JHI_St_60k_v1 | DMT400064456 | TGACACTCTTGTGTTCAAGTACATGAAGGGATCAAACTCAGTATTAGTGGTAAACAAGGA |
Microarray Signals from CUST_43302_PI426222305
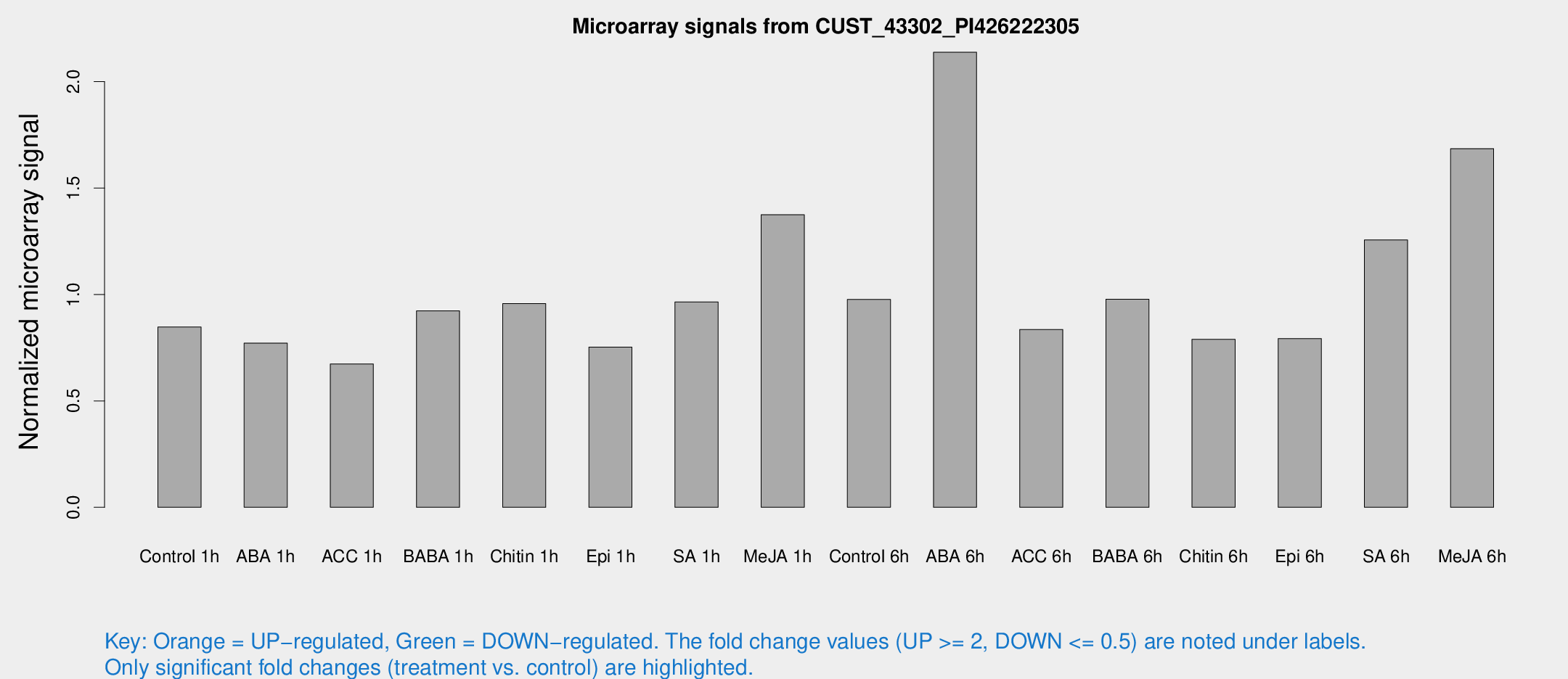
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 221.776 | 41.641 | 0.847217 | 0.101228 |
| ABA 1h | 178.807 | 31.7145 | 0.771191 | 0.121223 |
| ACC 1h | 195.216 | 62.7319 | 0.673484 | 0.196924 |
| BABA 1h | 244.186 | 60.9318 | 0.922934 | 0.188028 |
| Chitin 1h | 222.83 | 35.7278 | 0.956804 | 0.0889616 |
| Epi 1h | 168.935 | 25.8414 | 0.753029 | 0.11753 |
| SA 1h | 259.777 | 46.3904 | 0.964993 | 0.130116 |
| Me-JA 1h | 295.556 | 54.6247 | 1.37445 | 0.203271 |
| Control 6h | 262.586 | 59.9107 | 0.976625 | 0.170098 |
| ABA 6h | 633.935 | 171.334 | 2.13826 | 0.650258 |
| ACC 6h | 243.126 | 14.8708 | 0.835615 | 0.116932 |
| BABA 6h | 285.291 | 49.3819 | 0.977843 | 0.133366 |
| Chitin 6h | 215.194 | 24.4859 | 0.789174 | 0.0768759 |
| Epi 6h | 243.001 | 60.4192 | 0.792738 | 0.186684 |
| SA 6h | 320.498 | 42.9229 | 1.25641 | 0.0745427 |
| Me-JA 6h | 434.799 | 65.7961 | 1.68512 | 0.122024 |
Source Transcript PGSC0003DMT400064456 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G53870.1 | +2 | 1e-35 | 135 | 64/120 (53%) | early nodulin-like protein 1 | chr5:21870033-21871228 REVERSE LENGTH=370 |
