Probe CUST_43097_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43097_PI426222305 | JHI_St_60k_v1 | DMT400045724 | CATGGTCCACAAATTTTGCTAGAAAGATCTGATTCTTCAAGTCTTATGAGACATAGGTTA |
All Microarray Probes Designed to Gene DMG400017732
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_43097_PI426222305 | JHI_St_60k_v1 | DMT400045724 | CATGGTCCACAAATTTTGCTAGAAAGATCTGATTCTTCAAGTCTTATGAGACATAGGTTA |
| CUST_43078_PI426222305 | JHI_St_60k_v1 | DMT400045725 | GTACTGTACCTGATATATGTGCATGTGCAGGCAAAGTATAAGGCAAGAACAATTATAATT |
Microarray Signals from CUST_43097_PI426222305
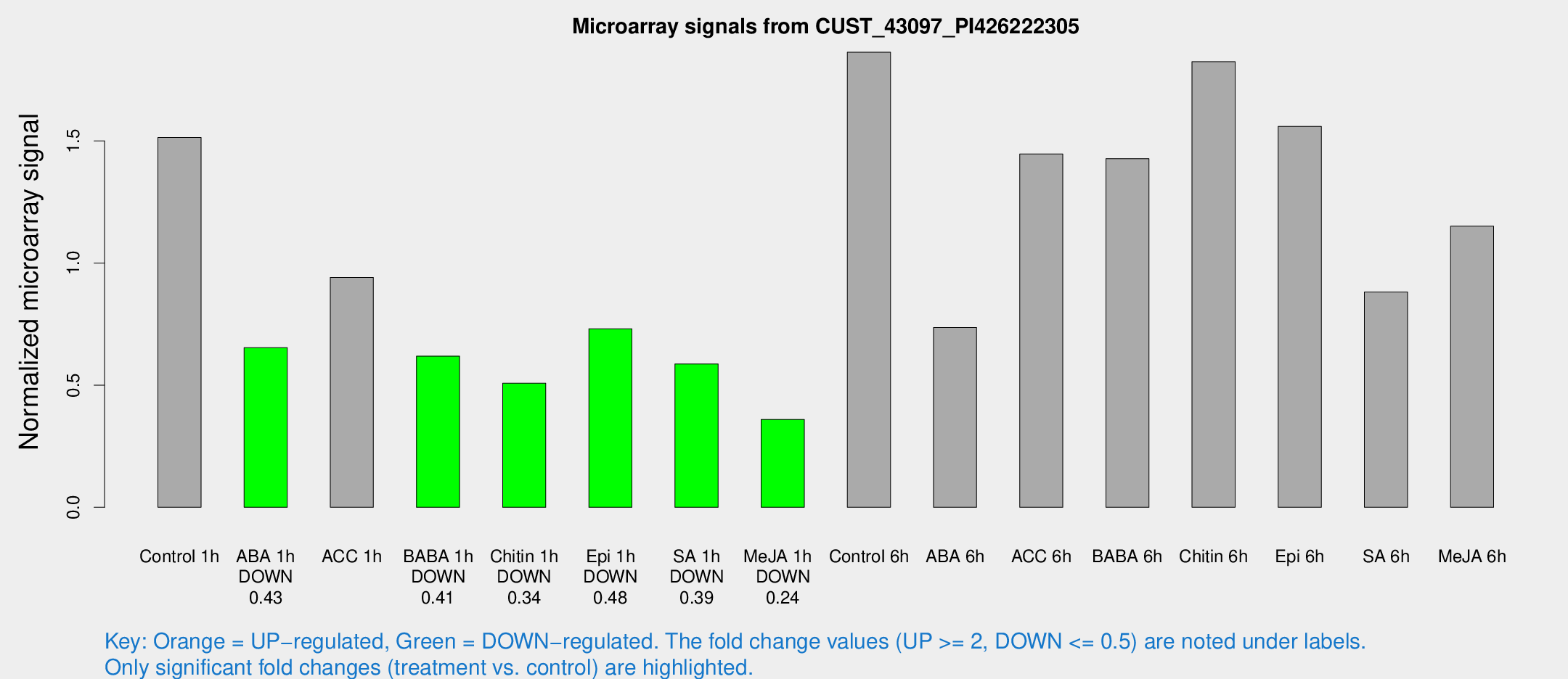
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1342.37 | 267.129 | 1.51398 | 0.222073 |
| ABA 1h | 501.081 | 67.0908 | 0.653385 | 0.0643764 |
| ACC 1h | 884.053 | 214.204 | 0.941299 | 0.199936 |
| BABA 1h | 555.089 | 144.559 | 0.618827 | 0.138186 |
| Chitin 1h | 391.649 | 37.2747 | 0.507855 | 0.0296284 |
| Epi 1h | 538.012 | 31.2624 | 0.730798 | 0.0424252 |
| SA 1h | 523.379 | 77.6016 | 0.586711 | 0.0502249 |
| Me-JA 1h | 259.806 | 50.1659 | 0.359901 | 0.0424826 |
| Control 6h | 1749.1 | 525.936 | 1.86316 | 0.481105 |
| ABA 6h | 667.48 | 41.0919 | 0.736032 | 0.0426689 |
| ACC 6h | 1495.98 | 360.395 | 1.44644 | 0.183036 |
| BABA 6h | 1405.7 | 272.508 | 1.42742 | 0.251894 |
| Chitin 6h | 1675.69 | 212.203 | 1.82465 | 0.181761 |
| Epi 6h | 1517.09 | 193.299 | 1.55957 | 0.208862 |
| SA 6h | 874.809 | 286.629 | 0.881557 | 0.3153 |
| Me-JA 6h | 1017.4 | 206.986 | 1.15079 | 0.147751 |
Source Transcript PGSC0003DMT400045724 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G12750.1 | +3 | 6e-96 | 296 | 171/335 (51%) | zinc transporter 1 precursor | chr3:4051950-4053156 REVERSE LENGTH=355 |
