Probe CUST_42739_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42739_PI426222305 | JHI_St_60k_v1 | DMT400052363 | GACATGTCTTAAGATATCAGCCATGGTATCCTGATCTTATTGTTTACTTCCAATGCTAAA |
All Microarray Probes Designed to Gene DMG400020326
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42739_PI426222305 | JHI_St_60k_v1 | DMT400052363 | GACATGTCTTAAGATATCAGCCATGGTATCCTGATCTTATTGTTTACTTCCAATGCTAAA |
| CUST_42728_PI426222305 | JHI_St_60k_v1 | DMT400052362 | AAGTCGTATCCAGCTTCTTCTGTTAATGCACATTCCCCCGCTTCATCTGTAGCAGAATCA |
Microarray Signals from CUST_42739_PI426222305
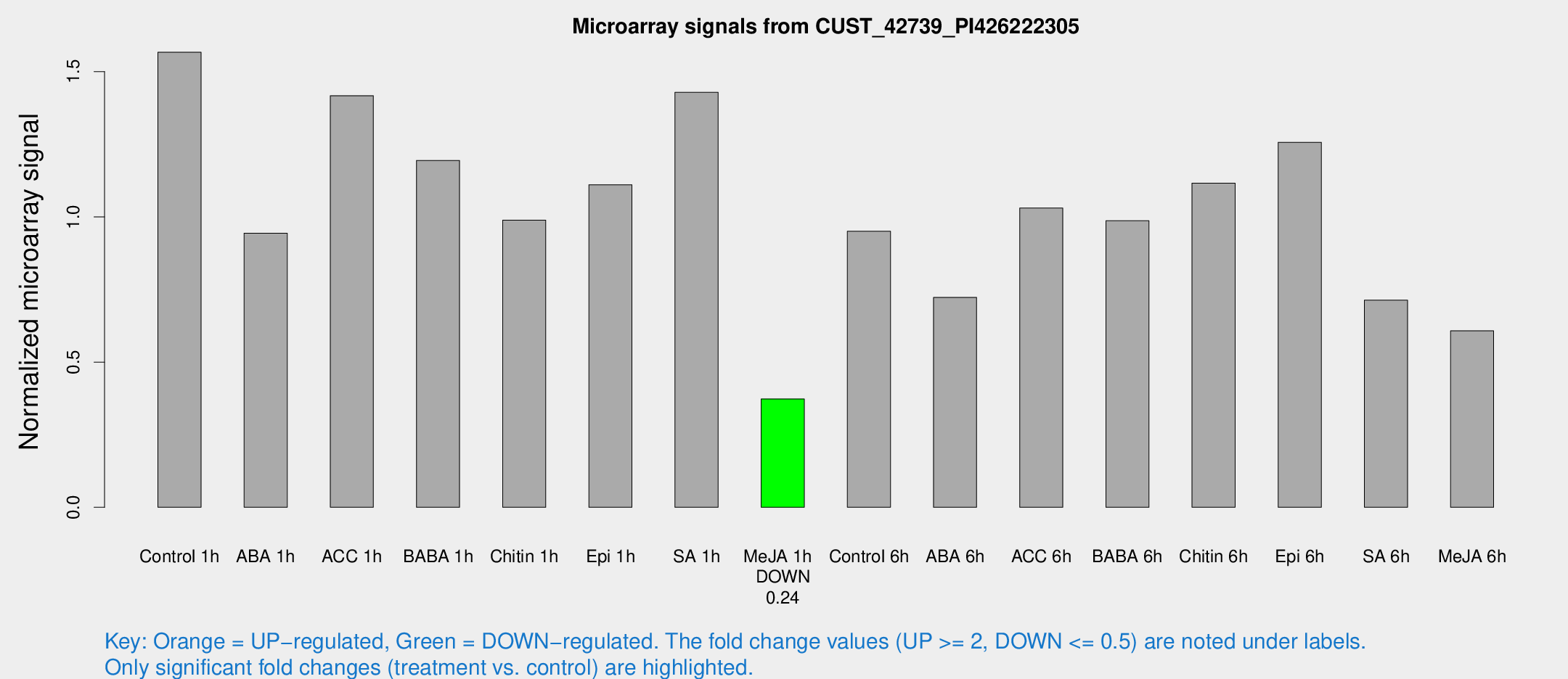
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 626.501 | 88.7462 | 1.56698 | 0.114747 |
| ABA 1h | 330.096 | 28.9669 | 0.944051 | 0.0552392 |
| ACC 1h | 605.2 | 132.028 | 1.41735 | 0.252494 |
| BABA 1h | 462.37 | 71.4723 | 1.19447 | 0.100802 |
| Chitin 1h | 351.504 | 32.0267 | 0.988888 | 0.0951053 |
| Epi 1h | 379.372 | 30.5038 | 1.11081 | 0.0923584 |
| SA 1h | 582.506 | 65.956 | 1.42876 | 0.10383 |
| Me-JA 1h | 121.84 | 16.5126 | 0.373268 | 0.0526601 |
| Control 6h | 411.142 | 116.08 | 0.950579 | 0.226277 |
| ABA 6h | 303.596 | 26.6177 | 0.722815 | 0.0425292 |
| ACC 6h | 472.43 | 62.5434 | 1.03055 | 0.131133 |
| BABA 6h | 437.314 | 43.7617 | 0.986577 | 0.060922 |
| Chitin 6h | 469.929 | 43.243 | 1.11594 | 0.0875779 |
| Epi 6h | 569.979 | 92.2703 | 1.25692 | 0.248445 |
| SA 6h | 288.167 | 55.0706 | 0.71324 | 0.0725368 |
| Me-JA 6h | 258.402 | 76.2715 | 0.607557 | 0.130839 |
Source Transcript PGSC0003DMT400052363 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37710.1 | +1 | 0.0 | 760 | 401/666 (60%) | receptor lectin kinase | chr2:15814934-15816961 REVERSE LENGTH=675 |
