Probe CUST_42594_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42594_PI426222305 | JHI_St_60k_v1 | DMT400026852 | GGATTGCCTTCAGCAAGAAATCTAAATGTGGAACTTGATGATGATTTCCTTAATGTATCA |
All Microarray Probes Designed to Gene DMG400010362
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_42597_PI426222305 | JHI_St_60k_v1 | DMT400026850 | GGTGATGCTGTCCTTGATGCCTCAATGTAAAATTGATATTTCTTCAACAAGTACATAACA |
| CUST_42594_PI426222305 | JHI_St_60k_v1 | DMT400026852 | GGATTGCCTTCAGCAAGAAATCTAAATGTGGAACTTGATGATGATTTCCTTAATGTATCA |
Microarray Signals from CUST_42594_PI426222305
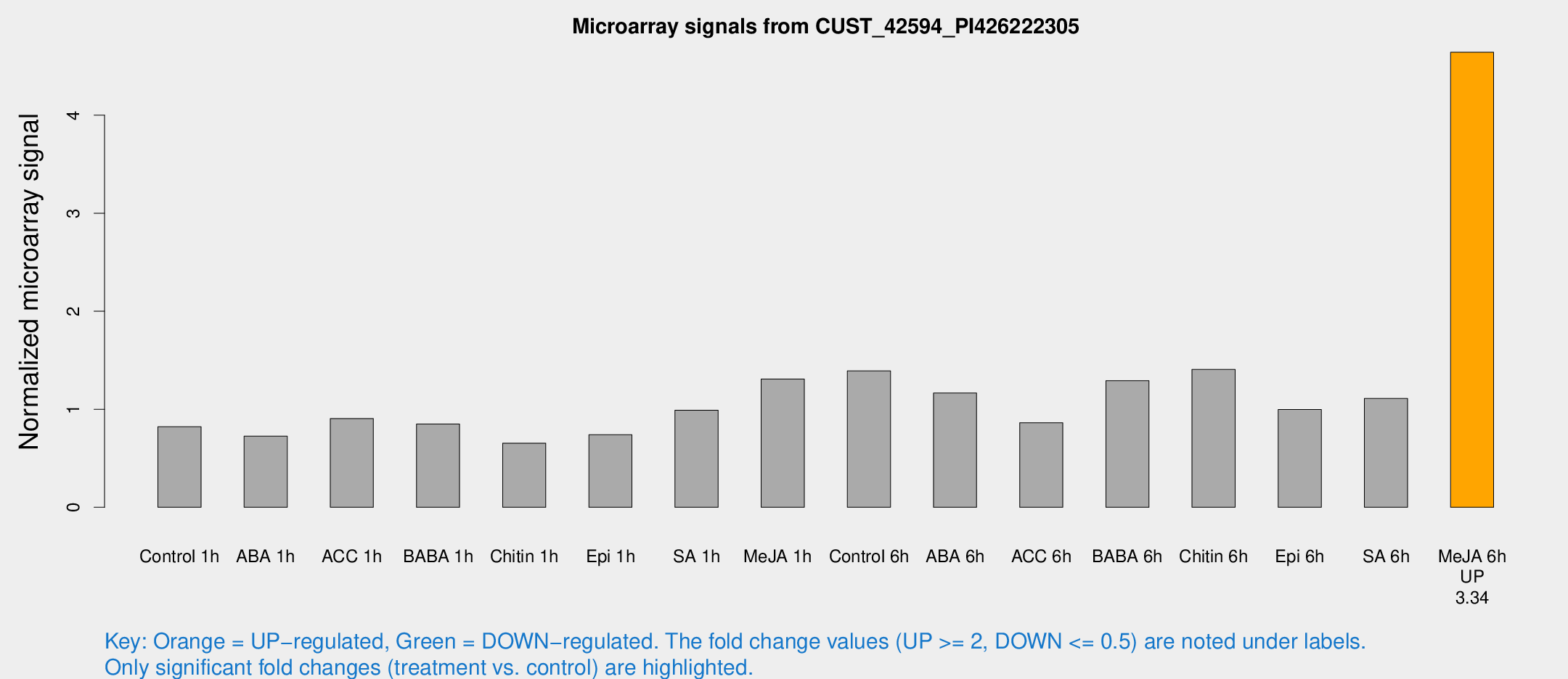
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 856.197 | 81.5083 | 0.821967 | 0.0475827 |
| ABA 1h | 684.297 | 120.941 | 0.726248 | 0.077289 |
| ACC 1h | 1024.5 | 237.296 | 0.904822 | 0.174899 |
| BABA 1h | 846.263 | 49.0739 | 0.849047 | 0.0659713 |
| Chitin 1h | 609.541 | 35.9496 | 0.654073 | 0.0379662 |
| Epi 1h | 671.196 | 77.7834 | 0.73994 | 0.0831521 |
| SA 1h | 1143.25 | 322.527 | 0.99087 | 0.354318 |
| Me-JA 1h | 1103.15 | 63.8005 | 1.3084 | 0.116313 |
| Control 6h | 1554.04 | 419.906 | 1.3918 | 0.273513 |
| ABA 6h | 1298.46 | 149.68 | 1.16559 | 0.148482 |
| ACC 6h | 1032.66 | 108.919 | 0.86253 | 0.0499395 |
| BABA 6h | 1493.86 | 86.3976 | 1.2916 | 0.0746619 |
| Chitin 6h | 1575.01 | 220.968 | 1.40649 | 0.204976 |
| Epi 6h | 1188.93 | 177.565 | 0.998309 | 0.235775 |
| SA 6h | 1183.16 | 223.199 | 1.11034 | 0.107049 |
| Me-JA 6h | 4844.28 | 579.901 | 4.64254 | 0.268064 |
Source Transcript PGSC0003DMT400026852 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G02300.1 | +1 | 8e-163 | 471 | 230/409 (56%) | Cysteine proteinases superfamily protein | chr1:453288-455376 FORWARD LENGTH=379 |
