Probe CUST_41581_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41581_PI426222305 | JHI_St_60k_v1 | DMT400021570 | GGTCACATTGATGCTCTTATTAACAATGCTGGCGTTAGAGAATTTATCAATTTCGGTTCG |
All Microarray Probes Designed to Gene DMG400008363
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41581_PI426222305 | JHI_St_60k_v1 | DMT400021570 | GGTCACATTGATGCTCTTATTAACAATGCTGGCGTTAGAGAATTTATCAATTTCGGTTCG |
| CUST_41483_PI426222305 | JHI_St_60k_v1 | DMT400021571 | CACTATAATAGTCAAATTTCCTTTGGAAGCATCCATAAGATATTTGGGTAAATGGCATCG |
Microarray Signals from CUST_41581_PI426222305
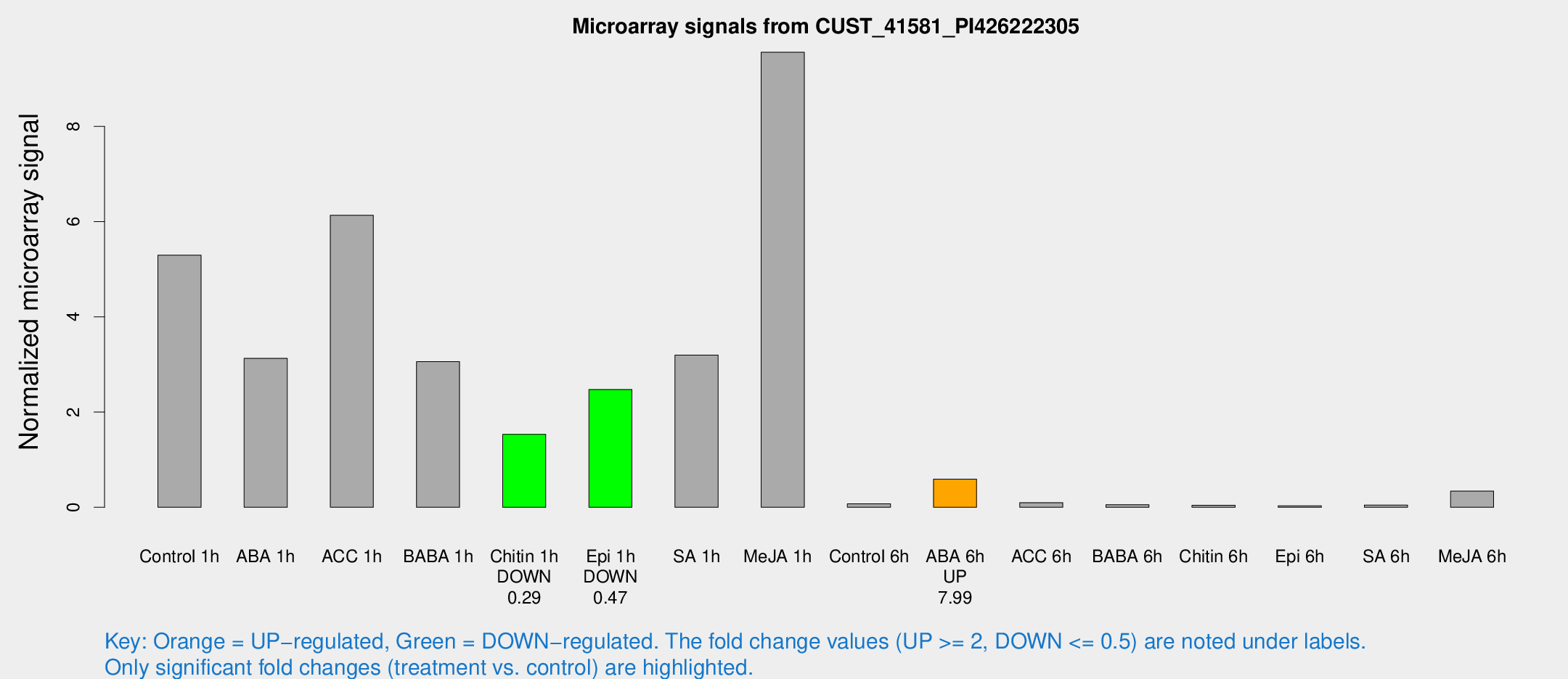
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2117.99 | 466.648 | 5.29688 | 0.815219 |
| ABA 1h | 1209.4 | 369.033 | 3.12977 | 1.19177 |
| ACC 1h | 2927.1 | 1166.81 | 6.13408 | 2.80065 |
| BABA 1h | 1175.15 | 248.537 | 3.05957 | 0.427679 |
| Chitin 1h | 542.08 | 90.3481 | 1.53252 | 0.208243 |
| Epi 1h | 827.722 | 97.6167 | 2.47406 | 0.292668 |
| SA 1h | 1484.8 | 606.928 | 3.19868 | 1.64103 |
| Me-JA 1h | 3072.51 | 573.224 | 9.55774 | 0.940928 |
| Control 6h | 35.8003 | 14.3358 | 0.0740181 | 0.0376454 |
| ABA 6h | 245.107 | 38.2901 | 0.591263 | 0.0617513 |
| ACC 6h | 57.5595 | 28.0778 | 0.0974576 | 0.0799982 |
| BABA 6h | 31.4973 | 14.4871 | 0.055077 | 0.0384952 |
| Chitin 6h | 17.8373 | 3.87238 | 0.0419969 | 0.0111367 |
| Epi 6h | 13.7612 | 4.07841 | 0.0311244 | 0.00987263 |
| SA 6h | 20.0165 | 7.07427 | 0.0468892 | 0.025869 |
| Me-JA 6h | 145.424 | 52.4901 | 0.340105 | 0.15875 |
Source Transcript PGSC0003DMT400021570 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G55310.1 | +1 | 1e-39 | 137 | 64/108 (59%) | NAD(P)-binding Rossmann-fold superfamily protein | chr3:20505873-20506958 FORWARD LENGTH=279 |
