Probe CUST_41094_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41094_PI426222305 | JHI_St_60k_v1 | DMT400004446 | CTCGTGGTTGCATTTGTTTAGTTTTCTTGTTTTTTGTCTGTGTTGCATCTTCAAGTGAAA |
All Microarray Probes Designed to Gene DMG400001774
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_41094_PI426222305 | JHI_St_60k_v1 | DMT400004446 | CTCGTGGTTGCATTTGTTTAGTTTTCTTGTTTTTTGTCTGTGTTGCATCTTCAAGTGAAA |
| CUST_41114_PI426222305 | JHI_St_60k_v1 | DMT400004447 | CTCGTGGTTGCATTTGTTTAGTTTTCTTGTTTTTTGTCTGTGTTGCATCTTCAAGTGAAA |
Microarray Signals from CUST_41094_PI426222305
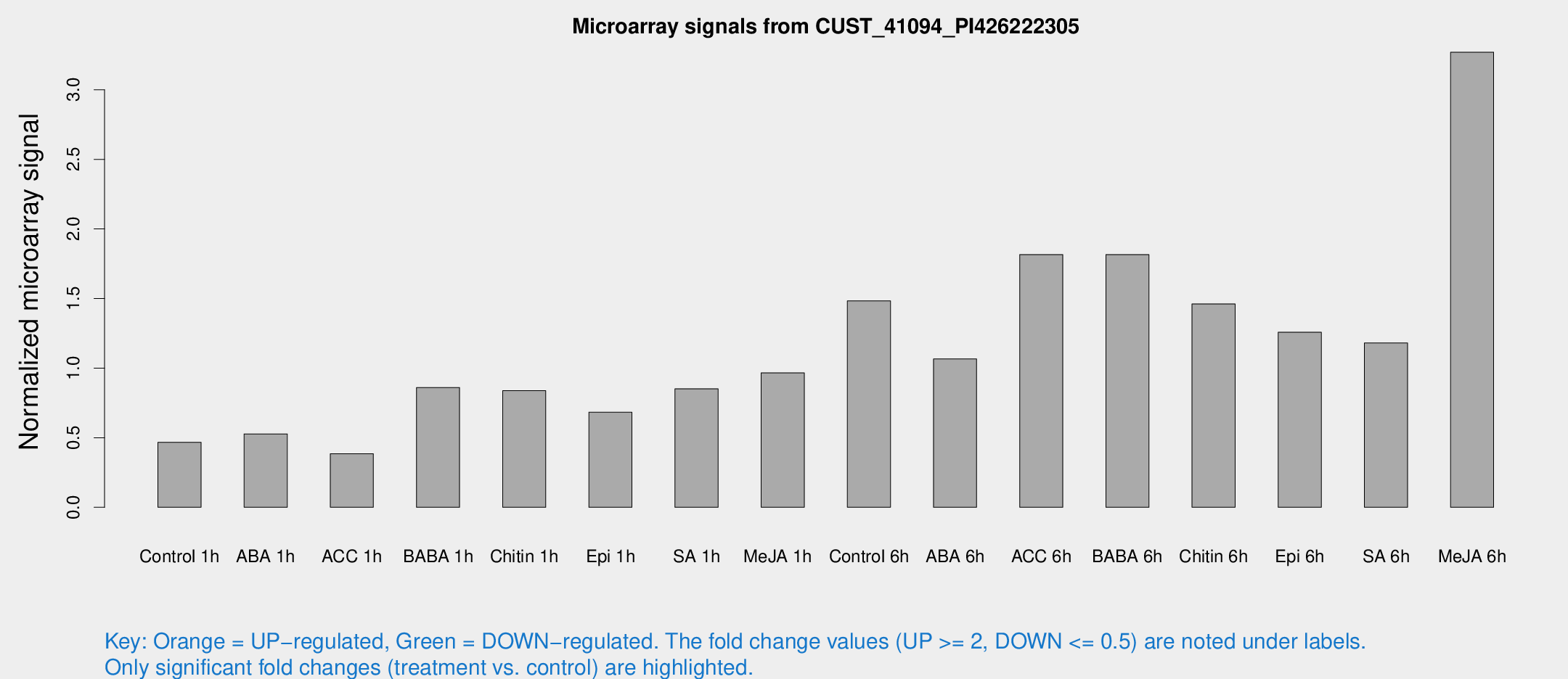
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 467.135 | 27.1995 | 0.46747 | 0.0271512 |
| ABA 1h | 497.293 | 115.554 | 0.527464 | 0.123124 |
| ACC 1h | 461.62 | 163.013 | 0.384384 | 0.149899 |
| BABA 1h | 864.49 | 170.546 | 0.86052 | 0.101692 |
| Chitin 1h | 784.118 | 153.699 | 0.838201 | 0.174983 |
| Epi 1h | 606.025 | 97.7905 | 0.682964 | 0.114082 |
| SA 1h | 883.333 | 103.281 | 0.851699 | 0.0556731 |
| Me-JA 1h | 830.807 | 206.104 | 0.965561 | 0.154455 |
| Control 6h | 1542.61 | 286.201 | 1.4837 | 0.179372 |
| ABA 6h | 1158.03 | 187.632 | 1.06581 | 0.10926 |
| ACC 6h | 2151.38 | 373.99 | 1.81589 | 0.371262 |
| BABA 6h | 2130.69 | 487.088 | 1.81574 | 0.344637 |
| Chitin 6h | 1570.77 | 174.813 | 1.46146 | 0.145992 |
| Epi 6h | 1430.43 | 158.951 | 1.25797 | 0.172309 |
| SA 6h | 1234.12 | 273.983 | 1.18054 | 0.162722 |
| Me-JA 6h | 3257.82 | 199.941 | 3.26967 | 0.303398 |
Source Transcript PGSC0003DMT400004446 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G37130.1 | +2 | 2e-162 | 463 | 218/300 (73%) | Peroxidase superfamily protein | chr2:15598225-15600004 REVERSE LENGTH=327 |
