Probe CUST_40747_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40747_PI426222305 | JHI_St_60k_v1 | DMT400073540 | CGGCGCAGAGAGAGAGAAGAGAAGAATCAGTTACCTAAATTTCTGAAATGAATACAGTAT |
All Microarray Probes Designed to Gene DMG400028568
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40747_PI426222305 | JHI_St_60k_v1 | DMT400073540 | CGGCGCAGAGAGAGAGAAGAGAAGAATCAGTTACCTAAATTTCTGAAATGAATACAGTAT |
Microarray Signals from CUST_40747_PI426222305
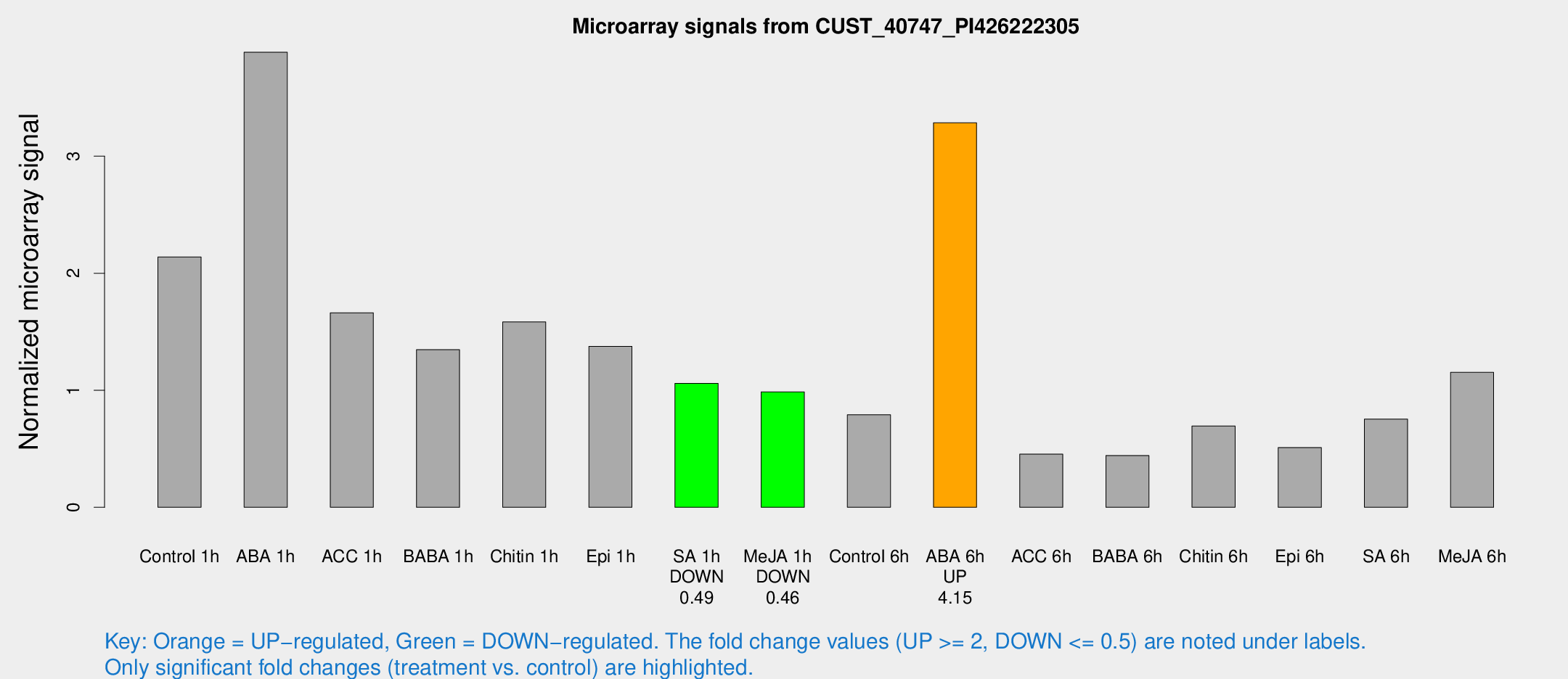
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1396.64 | 102.112 | 2.13887 | 0.123578 |
| ABA 1h | 2271.53 | 295.31 | 3.88816 | 0.486657 |
| ACC 1h | 1137.87 | 178.961 | 1.6619 | 0.171737 |
| BABA 1h | 902.355 | 209.288 | 1.34667 | 0.232432 |
| Chitin 1h | 947.923 | 153.744 | 1.58372 | 0.151668 |
| Epi 1h | 798.706 | 136.988 | 1.37546 | 0.247147 |
| SA 1h | 721.011 | 105.941 | 1.05848 | 0.088765 |
| Me-JA 1h | 525.249 | 36.5863 | 0.98552 | 0.0571938 |
| Control 6h | 529.96 | 85.8478 | 0.790975 | 0.0764297 |
| ABA 6h | 2293.27 | 228.979 | 3.28525 | 0.189733 |
| ACC 6h | 345.272 | 47.743 | 0.453771 | 0.0720123 |
| BABA 6h | 336.924 | 68.0338 | 0.44214 | 0.11505 |
| Chitin 6h | 481.525 | 28.0633 | 0.695017 | 0.0404358 |
| Epi 6h | 374.786 | 21.976 | 0.51072 | 0.0492204 |
| SA 6h | 484.827 | 28.217 | 0.753634 | 0.067323 |
| Me-JA 6h | 811.489 | 243.908 | 1.15281 | 0.290377 |
Source Transcript PGSC0003DMT400073540 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G54740.1 | +3 | 6e-21 | 92 | 54/121 (45%) | Protein of unknown function (DUF3049) | chr1:20430839-20431738 FORWARD LENGTH=299 |
