Probe CUST_40276_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40276_PI426222305 | JHI_St_60k_v1 | DMT400036486 | GTTGGAAGGCAAAGCCATAGACCTTCTTCCTTAGATTTGGAAGATTATTTAAGGATTAAA |
All Microarray Probes Designed to Gene DMG400014054
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40276_PI426222305 | JHI_St_60k_v1 | DMT400036486 | GTTGGAAGGCAAAGCCATAGACCTTCTTCCTTAGATTTGGAAGATTATTTAAGGATTAAA |
| CUST_40282_PI426222305 | JHI_St_60k_v1 | DMT400036485 | GTTGGAAGGCAAAGCCATAGACCTTCTTCCTTAGATTTGGAAGATTATTTAAGGATTAAA |
Microarray Signals from CUST_40276_PI426222305
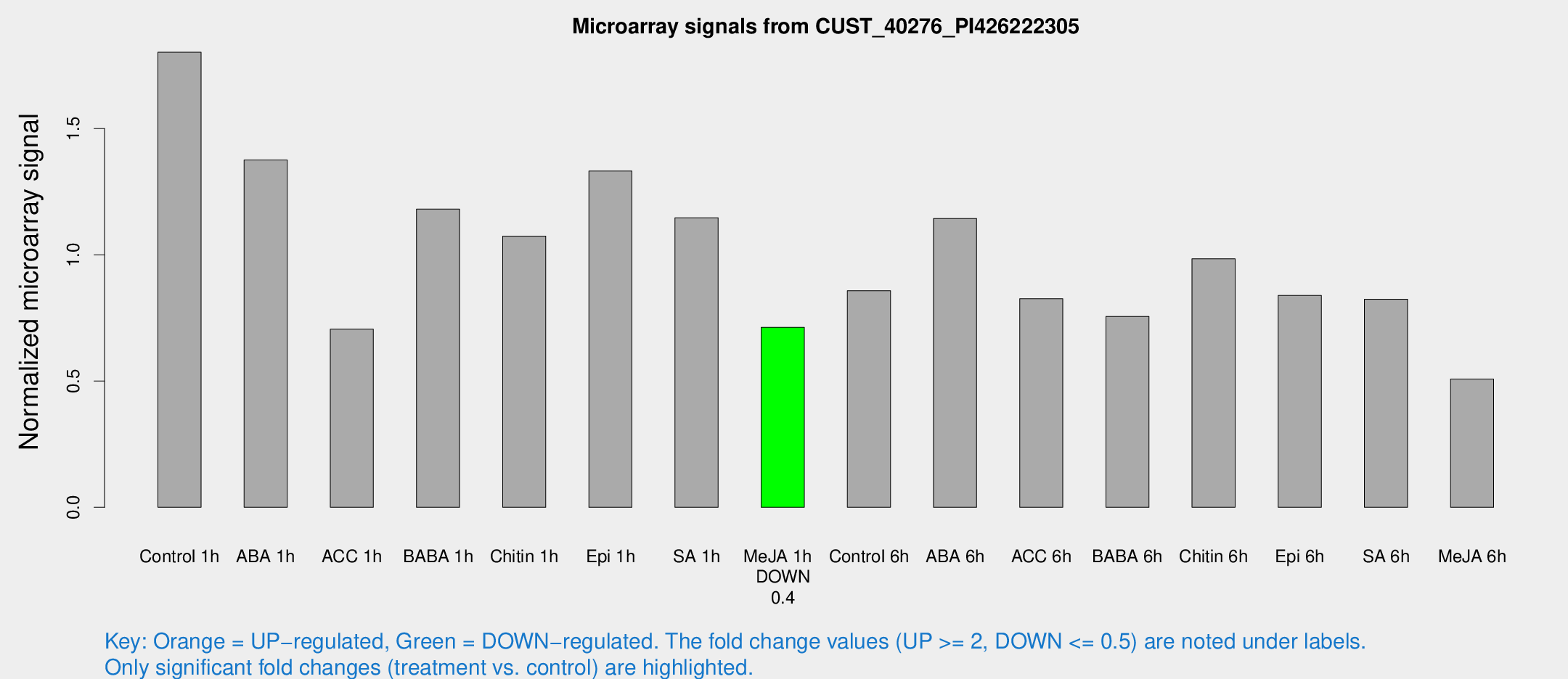
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2125.26 | 228.722 | 1.80273 | 0.104126 |
| ABA 1h | 1433.41 | 147.81 | 1.37566 | 0.0794938 |
| ACC 1h | 1011.33 | 348.32 | 0.70557 | 0.307161 |
| BABA 1h | 1402.76 | 317.817 | 1.18108 | 0.177673 |
| Chitin 1h | 1130.49 | 72.4537 | 1.07442 | 0.0621208 |
| Epi 1h | 1362.82 | 154.808 | 1.332 | 0.14385 |
| SA 1h | 1377.51 | 79.8503 | 1.14713 | 0.0662948 |
| Me-JA 1h | 678.748 | 39.3738 | 0.713003 | 0.0728114 |
| Control 6h | 1074.92 | 279.401 | 0.85826 | 0.177244 |
| ABA 6h | 1430.8 | 141.66 | 1.14405 | 0.06612 |
| ACC 6h | 1119.33 | 132.478 | 0.826222 | 0.047808 |
| BABA 6h | 1043.66 | 259.583 | 0.756174 | 0.174345 |
| Chitin 6h | 1221.76 | 70.6609 | 0.984605 | 0.0569347 |
| Epi 6h | 1106.03 | 64.1953 | 0.839131 | 0.0485526 |
| SA 6h | 962.531 | 107.326 | 0.824573 | 0.0485672 |
| Me-JA 6h | 638.623 | 184.752 | 0.508449 | 0.1035 |
Source Transcript PGSC0003DMT400036486 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G40460.1 | +3 | 6e-143 | 421 | 211/295 (72%) | Major facilitator superfamily protein | chr2:16897123-16901171 FORWARD LENGTH=583 |
