Probe CUST_40193_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40193_PI426222305 | JHI_St_60k_v1 | DMT400068388 | CAGGTTGAGTTGTATTCTGCTGCTAATTCTGCTTTTCAATACCATGTTATTTTCTACTTG |
All Microarray Probes Designed to Gene DMG400026594
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_40193_PI426222305 | JHI_St_60k_v1 | DMT400068388 | CAGGTTGAGTTGTATTCTGCTGCTAATTCTGCTTTTCAATACCATGTTATTTTCTACTTG |
Microarray Signals from CUST_40193_PI426222305
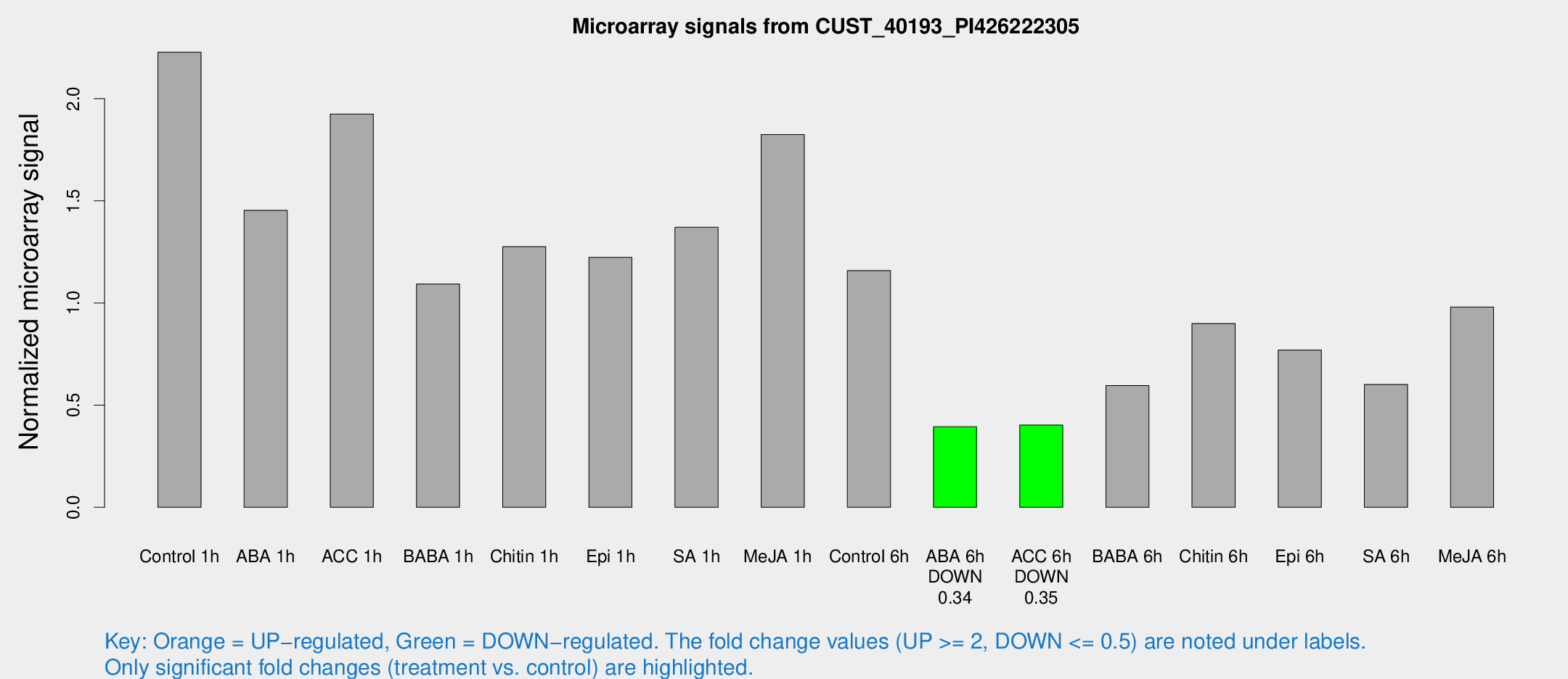
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 37750.7 | 2852.23 | 2.22744 | 0.316036 |
| ABA 1h | 23369.7 | 5687.7 | 1.45379 | 0.473164 |
| ACC 1h | 35203.4 | 7903.98 | 1.92484 | 0.322693 |
| BABA 1h | 17809.1 | 1031.43 | 1.09291 | 0.179825 |
| Chitin 1h | 20277.3 | 4513.41 | 1.27559 | 0.266773 |
| Epi 1h | 17910.7 | 1109.37 | 1.22312 | 0.0791532 |
| SA 1h | 28030.3 | 11520.6 | 1.3712 | 0.676627 |
| Me-JA 1h | 25896.7 | 4472.27 | 1.8241 | 0.458111 |
| Control 6h | 21356.3 | 5933.68 | 1.1584 | 0.254347 |
| ABA 6h | 7152 | 804.22 | 0.394406 | 0.0635836 |
| ACC 6h | 7922.73 | 1091.11 | 0.402484 | 0.0232383 |
| BABA 6h | 12236.9 | 3114.81 | 0.596331 | 0.218591 |
| Chitin 6h | 16164.1 | 934.329 | 0.899916 | 0.0519571 |
| Epi 6h | 15864.2 | 4153.05 | 0.769579 | 0.317382 |
| SA 6h | 10608.7 | 2546.83 | 0.601501 | 0.0983611 |
| Me-JA 6h | 20167.1 | 9295.6 | 0.980424 | 0.4355 |
Source Transcript PGSC0003DMT400068388 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G14690.1 | +1 | 5e-146 | 435 | 228/485 (47%) | cytochrome P450, family 72, subfamily A, polypeptide 15 | chr3:4937410-4939310 FORWARD LENGTH=512 |
