Probe CUST_39935_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39935_PI426222305 | JHI_St_60k_v1 | DMT400076978 | TTGACAAGAATTTCTGAATGCTGAATAATTGGACAGTGCTGCTTTCCGCGAAACCTTGCT |
All Microarray Probes Designed to Gene DMG400029941
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39941_PI426222305 | JHI_St_60k_v1 | DMT400076977 | CATTAGGTTCTGTGGCCAAATACCTTCAACTAGACAGAAAACAGGAAAAAGTGAATATTT |
| CUST_39935_PI426222305 | JHI_St_60k_v1 | DMT400076978 | TTGACAAGAATTTCTGAATGCTGAATAATTGGACAGTGCTGCTTTCCGCGAAACCTTGCT |
Microarray Signals from CUST_39935_PI426222305
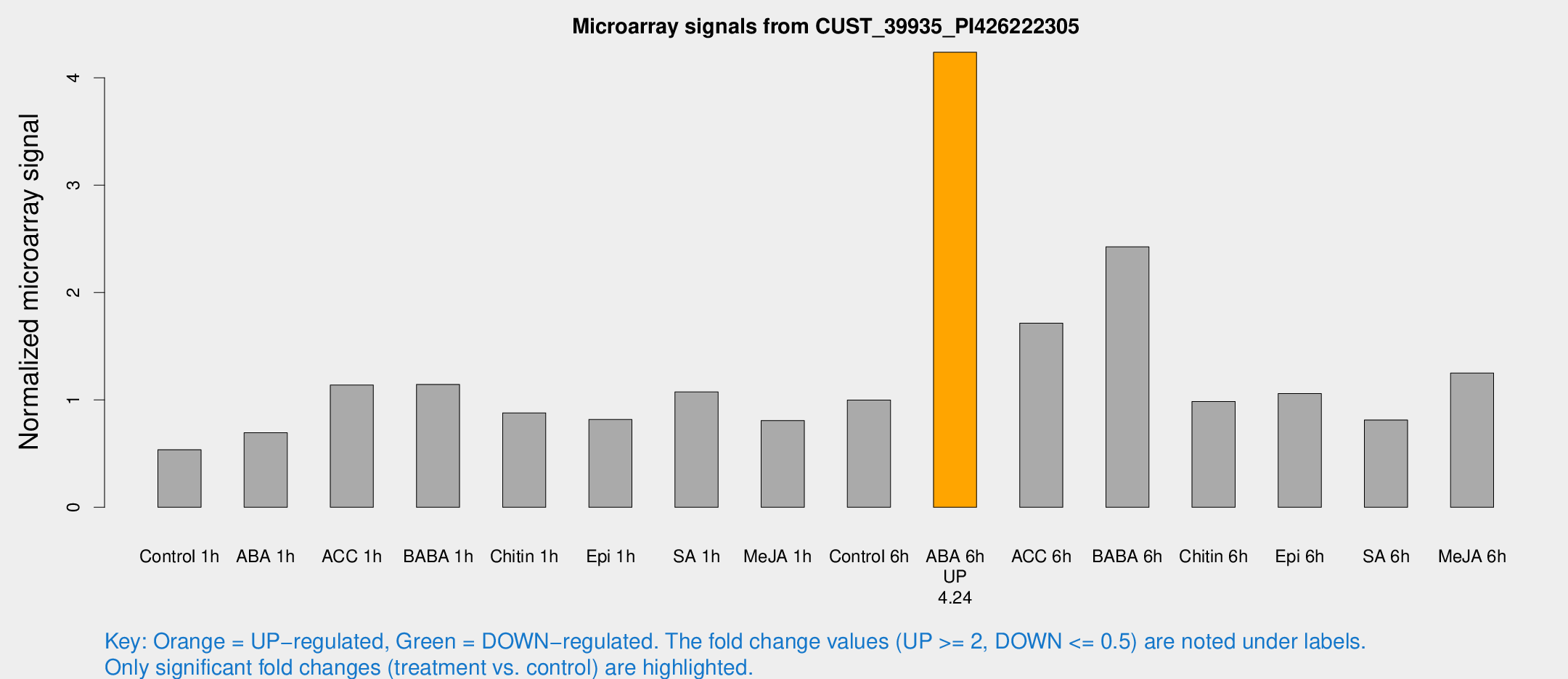
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 23.6415 | 5.1004 | 0.53637 | 0.119928 |
| ABA 1h | 26.6505 | 4.68734 | 0.694482 | 0.0975457 |
| ACC 1h | 50.6793 | 8.7293 | 1.1387 | 0.133258 |
| BABA 1h | 49.2608 | 12.6677 | 1.14454 | 0.209038 |
| Chitin 1h | 33.9446 | 5.40142 | 0.879403 | 0.194345 |
| Epi 1h | 30.7571 | 6.06951 | 0.818005 | 0.138411 |
| SA 1h | 46.3998 | 4.17655 | 1.07382 | 0.0969041 |
| Me-JA 1h | 28.1287 | 3.76673 | 0.808297 | 0.107254 |
| Control 6h | 46.7546 | 14.1441 | 0.998967 | 0.26258 |
| ABA 6h | 190.93 | 20.2828 | 4.2376 | 0.261287 |
| ACC 6h | 85.2567 | 15.6472 | 1.71411 | 0.214752 |
| BABA 6h | 121.205 | 31.3015 | 2.42589 | 0.588154 |
| Chitin 6h | 46.2552 | 10.8793 | 0.985762 | 0.223214 |
| Epi 6h | 51.3737 | 8.49348 | 1.05968 | 0.213143 |
| SA 6h | 34.866 | 6.01448 | 0.812928 | 0.103868 |
| Me-JA 6h | 58.8243 | 18.236 | 1.25062 | 0.41089 |
Source Transcript PGSC0003DMT400076978 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G26070.1 | +1 | 4e-52 | 178 | 83/139 (60%) | Protein of unknown function (DUF778) | chr2:11105741-11106607 REVERSE LENGTH=250 |
