Probe CUST_39328_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39328_PI426222305 | JHI_St_60k_v1 | DMT400012771 | TTAGCTTAGTTTGTGAGATTATGCAACTTGAGTTTCTTGATGAGAAGGATCGTGTGGGTC |
All Microarray Probes Designed to Gene DMG400004977
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_39320_PI426222305 | JHI_St_60k_v1 | DMT400012770 | ATTAAAGTCCGGTTTCTTCTTGTTGAATACAGTTTCTTGATGAGAAGGATCGTGTGGGTC |
| CUST_39328_PI426222305 | JHI_St_60k_v1 | DMT400012771 | TTAGCTTAGTTTGTGAGATTATGCAACTTGAGTTTCTTGATGAGAAGGATCGTGTGGGTC |
Microarray Signals from CUST_39328_PI426222305
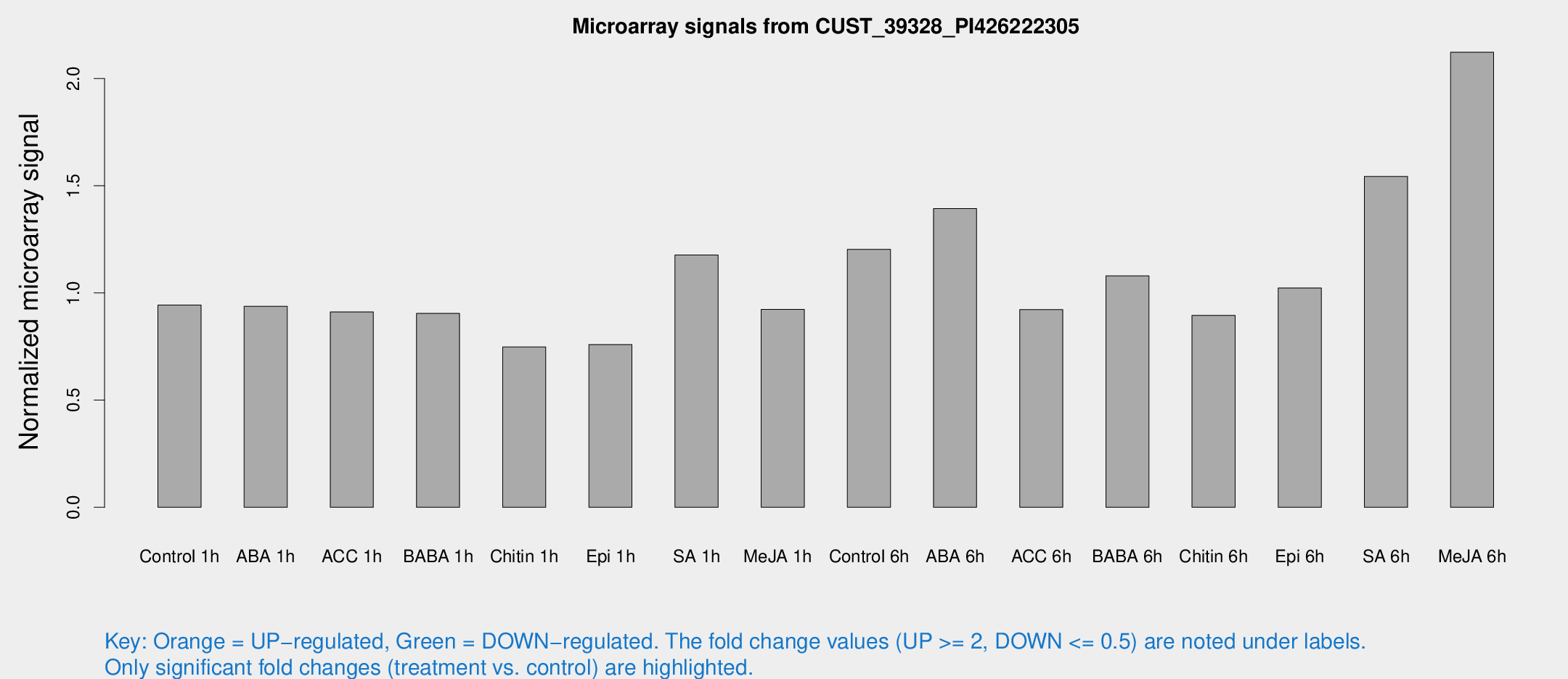
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5458.5 | 829.443 | 0.943528 | 0.0784542 |
| ABA 1h | 4758.56 | 554.375 | 0.937888 | 0.0541535 |
| ACC 1h | 5481.47 | 947.136 | 0.911188 | 0.104347 |
| BABA 1h | 5082.41 | 839.677 | 0.904482 | 0.0726033 |
| Chitin 1h | 3881.41 | 541.464 | 0.748085 | 0.04609 |
| Epi 1h | 3750.78 | 347.663 | 0.759044 | 0.0667562 |
| SA 1h | 6976.96 | 957.19 | 1.17684 | 0.133199 |
| Me-JA 1h | 4330.76 | 525.362 | 0.922929 | 0.0532914 |
| Control 6h | 7471.27 | 1966 | 1.20369 | 0.286282 |
| ABA 6h | 8452.54 | 778.128 | 1.39335 | 0.0804474 |
| ACC 6h | 6025.61 | 499.697 | 0.922357 | 0.0598148 |
| BABA 6h | 6846.75 | 395.726 | 1.08002 | 0.0644471 |
| Chitin 6h | 5421.87 | 393.706 | 0.895775 | 0.072475 |
| Epi 6h | 6549.74 | 407.38 | 1.02298 | 0.0956031 |
| SA 6h | 10031.6 | 3181.08 | 1.54391 | 0.500787 |
| Me-JA 6h | 12053.5 | 1075.87 | 2.12303 | 0.172277 |
Source Transcript PGSC0003DMT400012771 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G41210.1 | +1 | 1e-106 | 316 | 146/237 (62%) | glutathione S-transferase THETA 1 | chr5:16492550-16493884 REVERSE LENGTH=245 |
