Probe CUST_38955_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38955_PI426222305 | JHI_St_60k_v1 | DMT400070251 | TCTTTCGCATTTGAGTCTAGGAGTTTCTCATGACTTCCATGAATTGAATCATTTTGCTAG |
All Microarray Probes Designed to Gene DMG400027316
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38957_PI426222305 | JHI_St_60k_v1 | DMT400070252 | TCTTTCGCATTTGAGTCTAGGAGTTTCTCATGACTTCCATGAATTGAATCATTTTGCTAG |
| CUST_38955_PI426222305 | JHI_St_60k_v1 | DMT400070251 | TCTTTCGCATTTGAGTCTAGGAGTTTCTCATGACTTCCATGAATTGAATCATTTTGCTAG |
Microarray Signals from CUST_38955_PI426222305
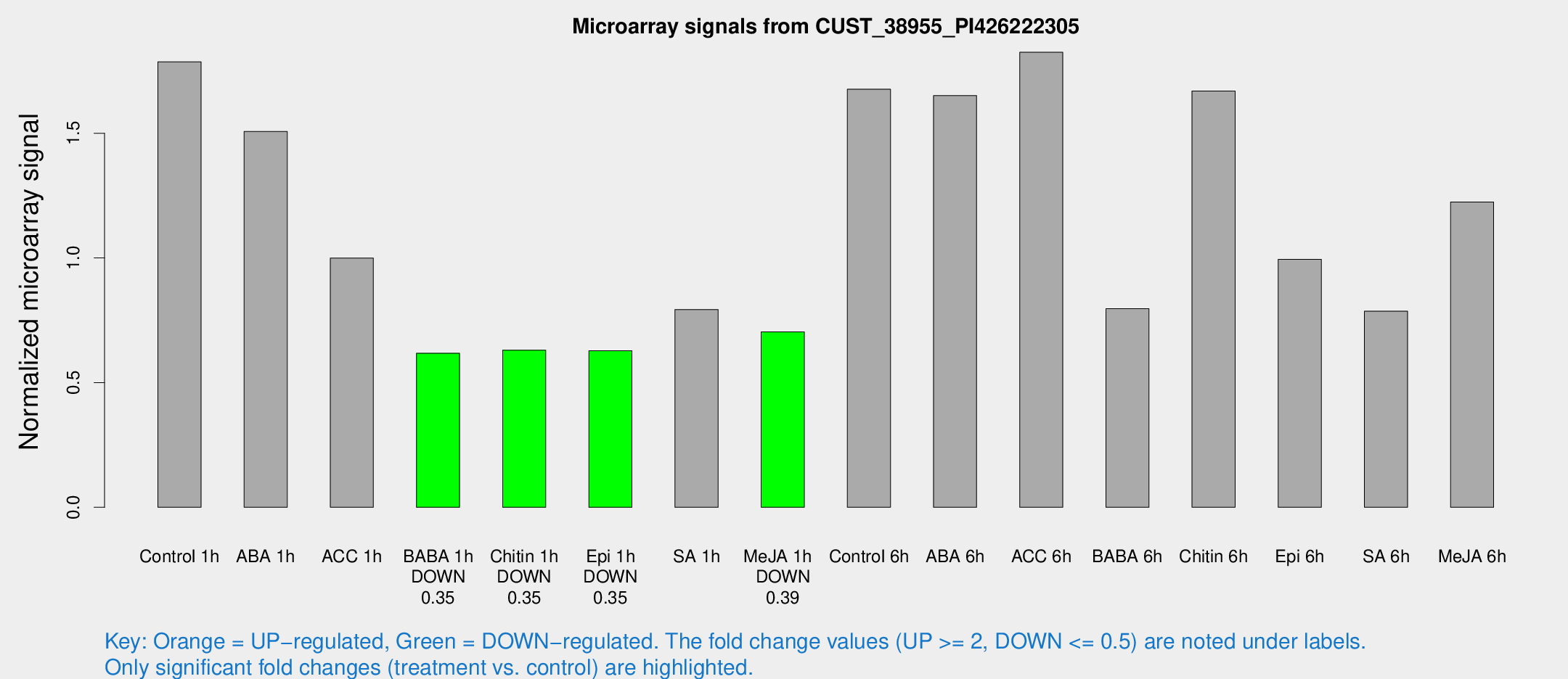
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 20.8423 | 3.88435 | 1.78686 | 0.347724 |
| ABA 1h | 15.5945 | 3.67477 | 1.50744 | 0.377322 |
| ACC 1h | 12.406 | 4.11704 | 0.999513 | 0.386868 |
| BABA 1h | 6.81373 | 3.96073 | 0.618059 | 0.35796 |
| Chitin 1h | 6.48658 | 3.77982 | 0.630235 | 0.365698 |
| Epi 1h | 6.21998 | 3.62298 | 0.627884 | 0.364247 |
| SA 1h | 10.8153 | 4.41982 | 0.793266 | 0.361824 |
| Me-JA 1h | 6.59 | 3.84188 | 0.703658 | 0.407427 |
| Control 6h | 23.6392 | 10.7445 | 1.67688 | 0.753374 |
| ABA 6h | 20.8408 | 4.14624 | 1.65088 | 0.360727 |
| ACC 6h | 27.5133 | 10.35 | 1.82506 | 0.815325 |
| BABA 6h | 10.9442 | 4.44657 | 0.796538 | 0.378808 |
| Chitin 6h | 21.3918 | 4.67871 | 1.6694 | 0.414551 |
| Epi 6h | 14.2541 | 4.94978 | 0.99464 | 0.403838 |
| SA 6h | 9.17921 | 4.19481 | 0.786546 | 0.38431 |
| Me-JA 6h | 16.2205 | 6.1271 | 1.22448 | 0.514227 |
Source Transcript PGSC0003DMT400070251 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G57810.2 | +2 | 2e-35 | 138 | 60/93 (65%) | Cysteine proteinases superfamily protein | chr3:21416336-21417809 FORWARD LENGTH=317 |
