Probe CUST_38444_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38444_PI426222305 | JHI_St_60k_v1 | DMT400074357 | CTGGTCTCTAAGATATCTCCACCATTTCCTCTGTATAATTCGTCACTATCTCTTTTTCGG |
All Microarray Probes Designed to Gene DMG401028902
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38383_PI426222305 | JHI_St_60k_v1 | DMT400074356 | CTGGTCTCTAAGATATCTCCACCATTTCCTCTGTATAATTCGTCACTATCTCTTTTTCGG |
| CUST_38444_PI426222305 | JHI_St_60k_v1 | DMT400074357 | CTGGTCTCTAAGATATCTCCACCATTTCCTCTGTATAATTCGTCACTATCTCTTTTTCGG |
Microarray Signals from CUST_38444_PI426222305
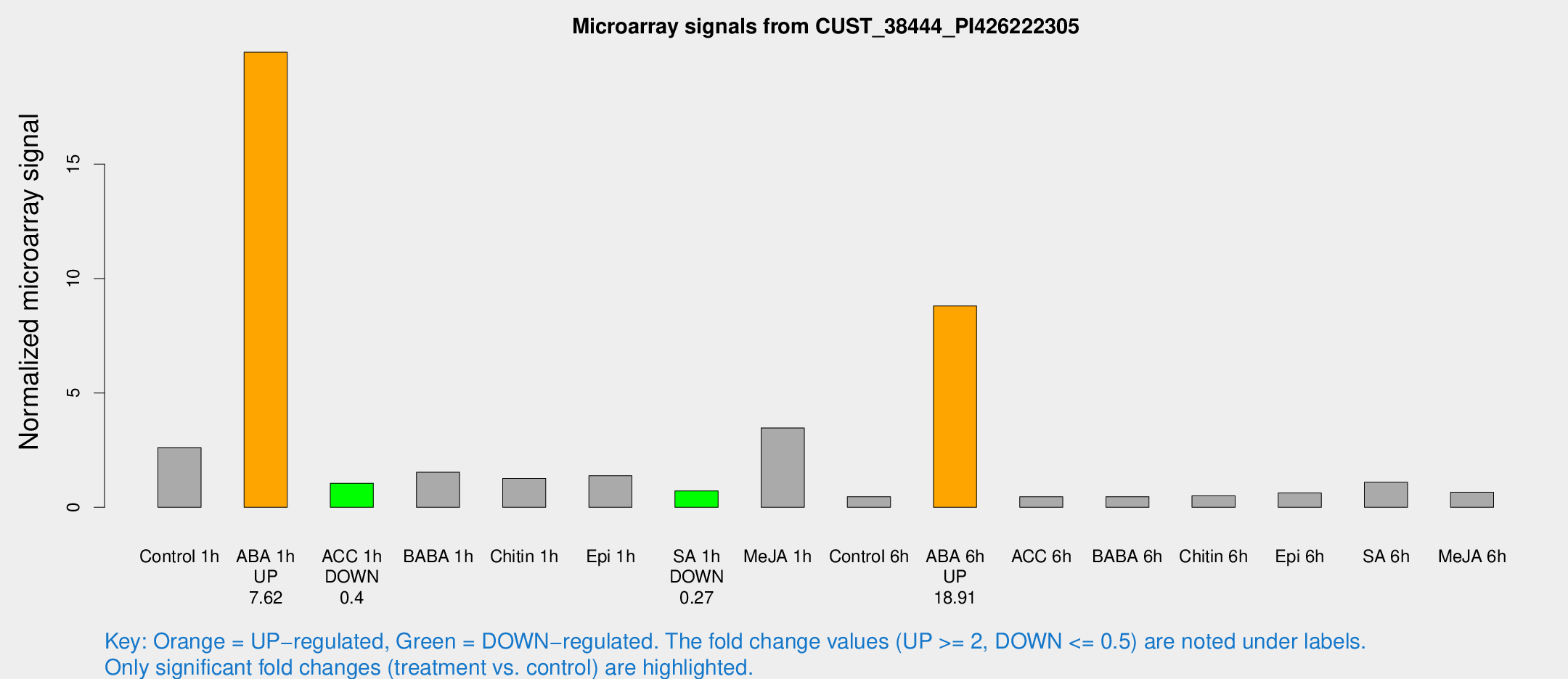
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 37.5383 | 4.75574 | 2.60944 | 0.282117 |
| ABA 1h | 250.456 | 17.256 | 19.8942 | 1.38311 |
| ACC 1h | 16.3762 | 4.51979 | 1.05114 | 0.368684 |
| BABA 1h | 21.8811 | 4.80318 | 1.53355 | 0.289542 |
| Chitin 1h | 17.2468 | 4.39253 | 1.26484 | 0.421947 |
| Epi 1h | 18.1723 | 5.0562 | 1.379 | 0.388779 |
| SA 1h | 10.8366 | 3.55965 | 0.71241 | 0.269914 |
| Me-JA 1h | 168.81 | 155.695 | 3.47076 | 29.5392 |
| Control 6h | 6.61063 | 3.80021 | 0.465395 | 0.267042 |
| ABA 6h | 141.54 | 33.0445 | 8.79937 | 2.13435 |
| ACC 6h | 7.62395 | 4.49944 | 0.461081 | 0.267369 |
| BABA 6h | 7.42448 | 4.3411 | 0.464874 | 0.269727 |
| Chitin 6h | 7.60648 | 4.42762 | 0.502297 | 0.291139 |
| Epi 6h | 10.8154 | 4.60529 | 0.63109 | 0.301339 |
| SA 6h | 16.1862 | 4.19325 | 1.09764 | 0.330603 |
| Me-JA 6h | 10.7735 | 4.38563 | 0.657463 | 0.309488 |
Source Transcript PGSC0003DMT400074357 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G24080.1 | +1 | 0.0 | 519 | 255/354 (72%) | Protein kinase superfamily protein | chr5:8139334-8141014 REVERSE LENGTH=470 |
