Probe CUST_38281_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38281_PI426222305 | JHI_St_60k_v1 | DMT400067343 | GACATTTACTTTGCCAGTACCTAGATGAGAGGATTTAGCTTTCCATATTTCTTATGGAAT |
All Microarray Probes Designed to Gene DMG400026183
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38260_PI426222305 | JHI_St_60k_v1 | DMT400067341 | GACATTTACTTTGCCAGTACCTAGATGAGAGGATTTAGCTTTCCATATTTCTTATGGAAT |
| CUST_38281_PI426222305 | JHI_St_60k_v1 | DMT400067343 | GACATTTACTTTGCCAGTACCTAGATGAGAGGATTTAGCTTTCCATATTTCTTATGGAAT |
Microarray Signals from CUST_38281_PI426222305
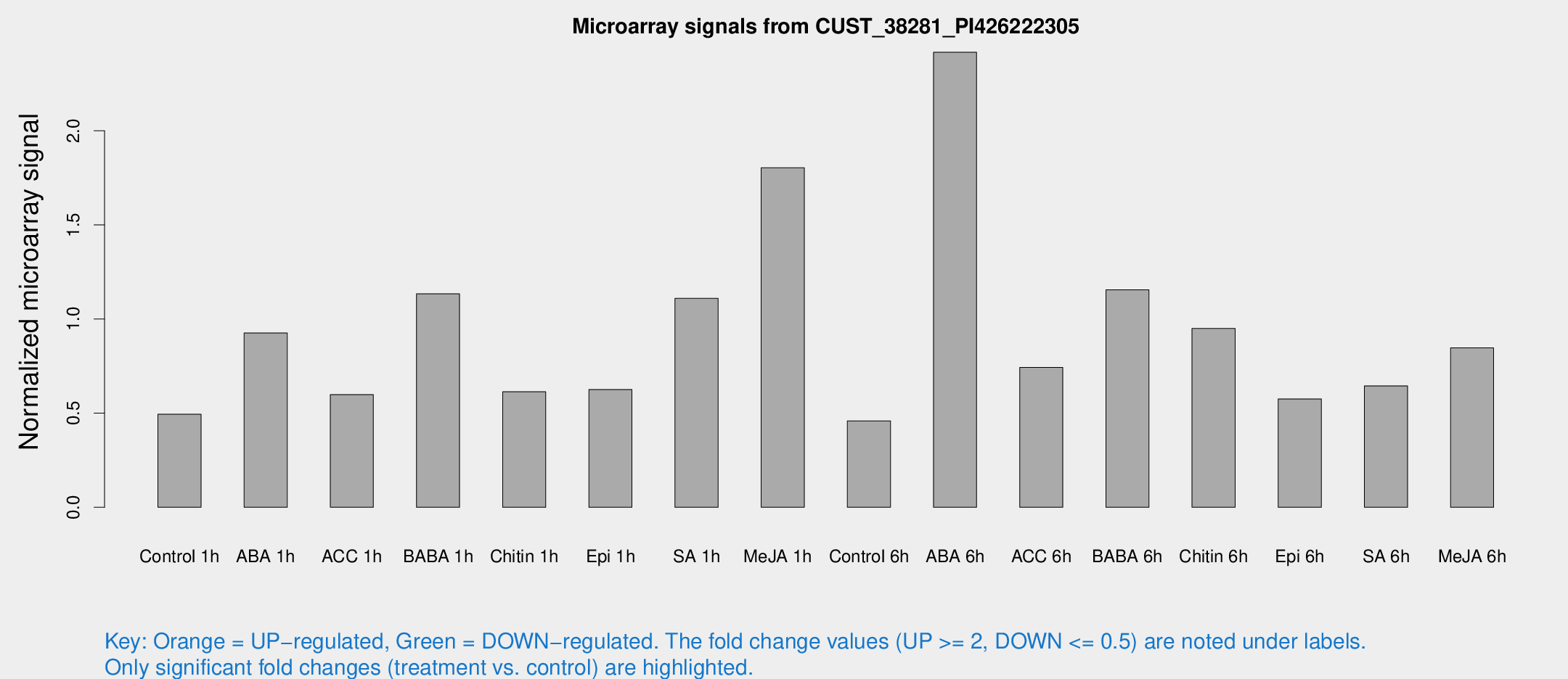
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 12.493 | 6.44132 | 0.494032 | 0.284901 |
| ABA 1h | 20.0286 | 7.24971 | 0.925928 | 0.512567 |
| ACC 1h | 14.1246 | 5.48518 | 0.598709 | 0.272001 |
| BABA 1h | 25.2227 | 8.9195 | 1.13408 | 0.392936 |
| Chitin 1h | 13.3116 | 6.25215 | 0.613472 | 0.248811 |
| Epi 1h | 11.6473 | 3.5127 | 0.625815 | 0.237759 |
| SA 1h | 23.1559 | 4.09983 | 1.10986 | 0.197635 |
| Me-JA 1h | 31.8312 | 10.1206 | 1.80335 | 0.437714 |
| Control 6h | 9.98545 | 3.78822 | 0.459202 | 0.215799 |
| ABA 6h | 51.4381 | 6.53203 | 2.41719 | 0.332733 |
| ACC 6h | 19.5229 | 6.85353 | 0.743025 | 0.235874 |
| BABA 6h | 25.7761 | 4.41291 | 1.15514 | 0.200538 |
| Chitin 6h | 20.8766 | 4.3063 | 0.949646 | 0.253943 |
| Epi 6h | 13.7781 | 4.48701 | 0.575652 | 0.211448 |
| SA 6h | 13.888 | 4.06091 | 0.644993 | 0.318747 |
| Me-JA 6h | 20.0414 | 6.90593 | 0.846813 | 0.390346 |
Source Transcript PGSC0003DMT400067343 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G21390.1 | +2 | 1e-166 | 365 | 189/363 (52%) | S-locus lectin protein kinase family protein | chr4:11394458-11397474 REVERSE LENGTH=849 |
