Probe CUST_38195_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38195_PI426222305 | JHI_St_60k_v1 | DMT400054792 | ACATGATTCTTCTGAGGATCTGAGTAACATTGTTGATCTGTAGACGAAGACGGAGATGTA |
All Microarray Probes Designed to Gene DMG400021265
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_38119_PI426222305 | JHI_St_60k_v1 | DMT400054793 | ACATGATTCTTCTGAGGATCTGAGTAACATTGTTGATCTGTAGACGAAGACGGAGATGTA |
| CUST_38195_PI426222305 | JHI_St_60k_v1 | DMT400054792 | ACATGATTCTTCTGAGGATCTGAGTAACATTGTTGATCTGTAGACGAAGACGGAGATGTA |
Microarray Signals from CUST_38195_PI426222305
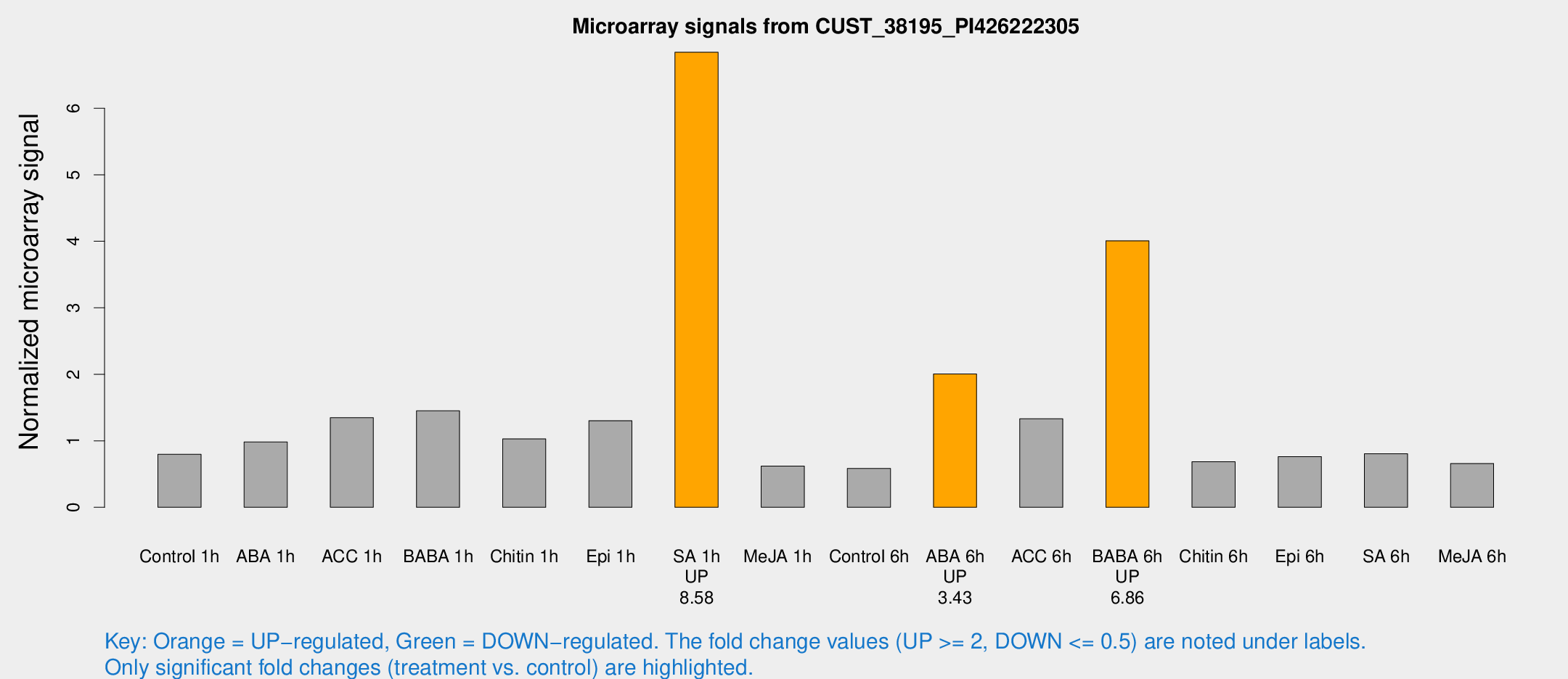
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 196.003 | 27.1231 | 0.797314 | 0.0643119 |
| ABA 1h | 211.695 | 19.2906 | 0.982405 | 0.153869 |
| ACC 1h | 367.956 | 98.6792 | 1.34755 | 0.355835 |
| BABA 1h | 344.199 | 47.6449 | 1.45108 | 0.113092 |
| Chitin 1h | 226.836 | 28.0059 | 1.02829 | 0.0618235 |
| Epi 1h | 272.627 | 16.1732 | 1.30205 | 0.0896997 |
| SA 1h | 1892.03 | 535.139 | 6.84279 | 2.23601 |
| Me-JA 1h | 124.138 | 14.2623 | 0.620633 | 0.0407243 |
| Control 6h | 154.957 | 45.3317 | 0.584393 | 0.132257 |
| ABA 6h | 567.525 | 170.652 | 2.00466 | 0.557235 |
| ACC 6h | 374.533 | 46.9476 | 1.33192 | 0.340119 |
| BABA 6h | 1121.52 | 206.55 | 4.00687 | 0.741228 |
| Chitin 6h | 176.32 | 11.0986 | 0.684453 | 0.0430154 |
| Epi 6h | 218.847 | 51.3622 | 0.762859 | 0.162127 |
| SA 6h | 192.47 | 11.8739 | 0.804242 | 0.0661038 |
| Me-JA 6h | 170.107 | 44.4183 | 0.657998 | 0.126886 |
Source Transcript PGSC0003DMT400054792 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G50930.1 | +1 | 2e-134 | 407 | 233/516 (45%) | cytochrome BC1 synthesis | chr3:18929817-18931547 FORWARD LENGTH=576 |
