Probe CUST_37778_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37778_PI426222305 | JHI_St_60k_v1 | DMT400077053 | CCTCCATTTGAACTATTGTGTACTTGACTAGCACATGTACTGCCTAGTATTGTTAGTGTC |
All Microarray Probes Designed to Gene DMG400029966
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37784_PI426222305 | JHI_St_60k_v1 | DMT400077052 | CTTGTTTACGCCTCGTTATAATGTTCTTATGGAAAATCACAAAGAGACATGAGTTTGAGT |
| CUST_37778_PI426222305 | JHI_St_60k_v1 | DMT400077053 | CCTCCATTTGAACTATTGTGTACTTGACTAGCACATGTACTGCCTAGTATTGTTAGTGTC |
Microarray Signals from CUST_37778_PI426222305
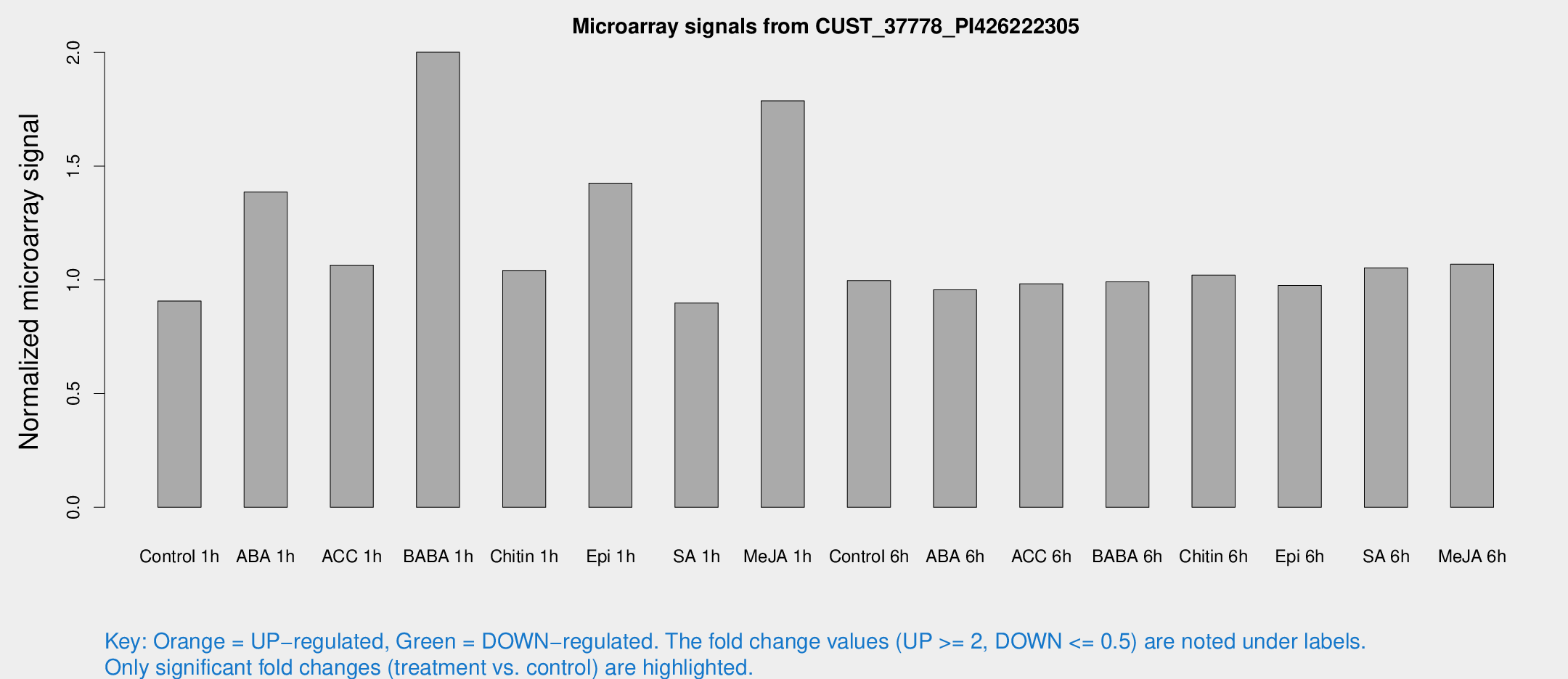
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.78976 | 3.36076 | 0.907248 | 0.526257 |
| ABA 1h | 8.5921 | 3.36305 | 1.38629 | 0.658245 |
| ACC 1h | 7.00811 | 3.74414 | 1.06529 | 0.574796 |
| BABA 1h | 19.3476 | 12.9169 | 2.00117 | 2.48275 |
| Chitin 1h | 6.00741 | 3.49851 | 1.04175 | 0.603265 |
| Epi 1h | 7.9787 | 3.39111 | 1.42555 | 0.629628 |
| SA 1h | 5.88934 | 3.42344 | 0.898079 | 0.521541 |
| Me-JA 1h | 11.0435 | 4.76219 | 1.78732 | 0.825126 |
| Control 6h | 6.37817 | 3.69722 | 0.997042 | 0.577384 |
| ABA 6h | 6.48982 | 3.76092 | 0.95661 | 0.553934 |
| ACC 6h | 7.35034 | 4.3553 | 0.983088 | 0.569387 |
| BABA 6h | 7.10529 | 4.13082 | 0.991673 | 0.574259 |
| Chitin 6h | 6.93108 | 4.01818 | 1.02075 | 0.591557 |
| Epi 6h | 7.0955 | 4.1568 | 0.975901 | 0.565109 |
| SA 6h | 6.63917 | 3.84556 | 1.05258 | 0.609642 |
| Me-JA 6h | 6.84371 | 3.73534 | 1.06885 | 0.588676 |
Source Transcript PGSC0003DMT400077053 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G10310.1 | +2 | 3e-57 | 201 | 106/217 (49%) | high-affinity K+ transporter 1 | chr4:6392008-6395667 FORWARD LENGTH=506 |
