Probe CUST_37657_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37657_PI426222305 | JHI_St_60k_v1 | DMT400049622 | CATCCTTGTCGATTGTGAGTGTCATTCATTTCTCAGAATTGTATTAAAATCCTCCCTTTT |
All Microarray Probes Designed to Gene DMG400019276
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37657_PI426222305 | JHI_St_60k_v1 | DMT400049622 | CATCCTTGTCGATTGTGAGTGTCATTCATTTCTCAGAATTGTATTAAAATCCTCCCTTTT |
| CUST_37587_PI426222305 | JHI_St_60k_v1 | DMT400049621 | AGACAATACAAGCCGTGTTGTTGGAACATATGGTTACATGTCCCCGGAATATGCAGTCGA |
Microarray Signals from CUST_37657_PI426222305
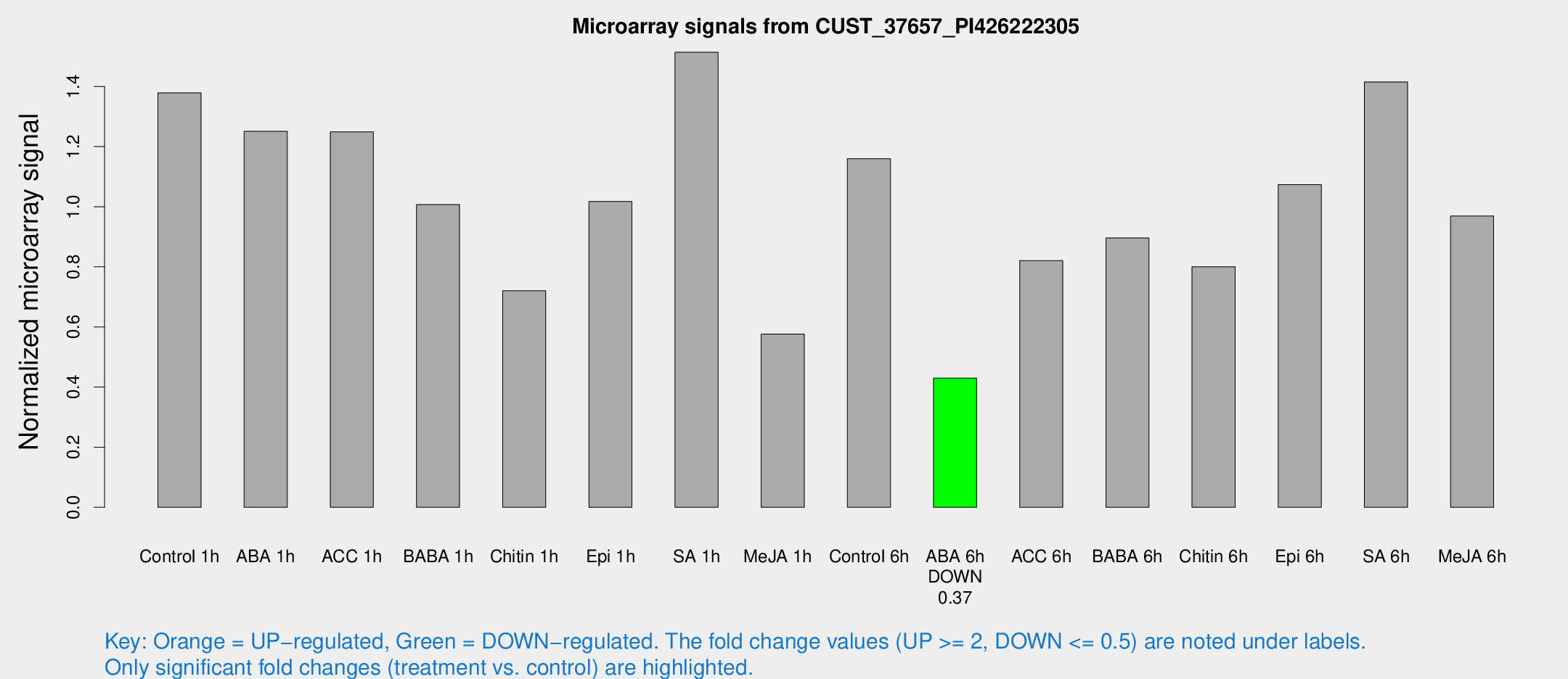
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 868.136 | 226.918 | 1.37874 | 0.262609 |
| ABA 1h | 667.529 | 93.4326 | 1.25058 | 0.165752 |
| ACC 1h | 826.303 | 226.532 | 1.24913 | 0.303176 |
| BABA 1h | 588.963 | 86.5779 | 1.00716 | 0.0584789 |
| Chitin 1h | 392.876 | 58.3305 | 0.720304 | 0.0637191 |
| Epi 1h | 522.76 | 30.4343 | 1.0174 | 0.0590807 |
| SA 1h | 936.025 | 114.917 | 1.5139 | 0.120388 |
| Me-JA 1h | 294.34 | 67.6516 | 0.576315 | 0.0884734 |
| Control 6h | 736.755 | 173.326 | 1.15953 | 0.220913 |
| ABA 6h | 274.137 | 30.7077 | 0.429965 | 0.0266109 |
| ACC 6h | 559.452 | 34.368 | 0.820654 | 0.102081 |
| BABA 6h | 596.813 | 43.7194 | 0.896036 | 0.0588902 |
| Chitin 6h | 513.687 | 72.0499 | 0.800412 | 0.0864419 |
| Epi 6h | 727.754 | 92.9332 | 1.07379 | 0.137687 |
| SA 6h | 842.034 | 109.745 | 1.41523 | 0.141719 |
| Me-JA 6h | 608.081 | 159.804 | 0.969097 | 0.177739 |
Source Transcript PGSC0003DMT400049622 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G21380.1 | +3 | 0.0 | 395 | 226/483 (47%) | receptor kinase 3 | chr4:11389219-11393090 REVERSE LENGTH=850 |
