Probe CUST_37640_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37640_PI426222305 | JHI_St_60k_v1 | DMT400049666 | CATGATTATTGAGGTGGAAAGGGCATTTATTCTTTGGTTGGCGACTAAAATAGCAAAAAA |
All Microarray Probes Designed to Gene DMG400019294
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37640_PI426222305 | JHI_St_60k_v1 | DMT400049666 | CATGATTATTGAGGTGGAAAGGGCATTTATTCTTTGGTTGGCGACTAAAATAGCAAAAAA |
| CUST_37647_PI426222305 | JHI_St_60k_v1 | DMT400049665 | CATGATTATTGAGGTGGAAAGGGCATTTATTCTTTGGTTGGCGACTAAAATAGCAAAAAA |
Microarray Signals from CUST_37640_PI426222305
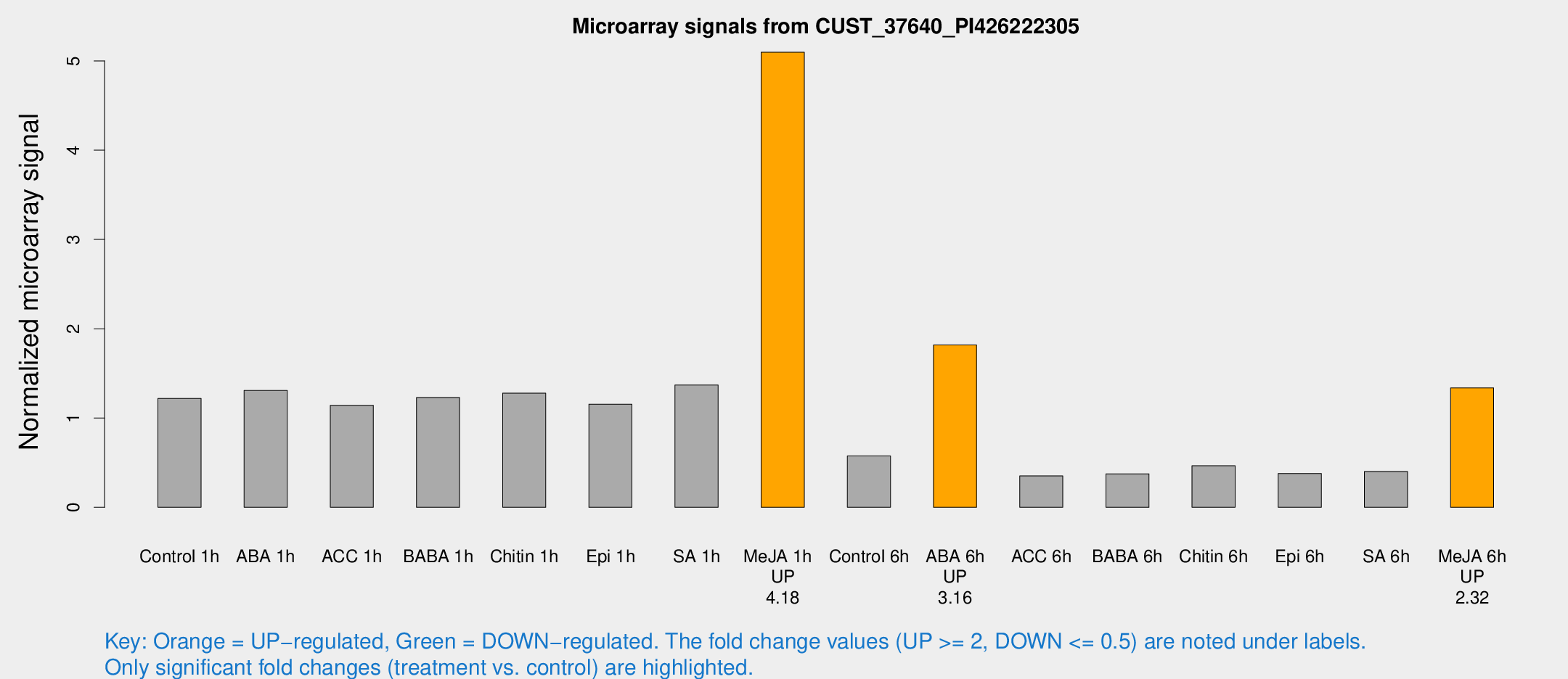
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5874.25 | 785.14 | 1.21974 | 0.167873 |
| ABA 1h | 5617.22 | 930.835 | 1.30815 | 0.123836 |
| ACC 1h | 6130 | 2038.84 | 1.14245 | 0.3327 |
| BABA 1h | 5857.41 | 1157.06 | 1.22976 | 0.145836 |
| Chitin 1h | 5448.68 | 315.556 | 1.27862 | 0.154339 |
| Epi 1h | 4826.72 | 741.858 | 1.15468 | 0.180784 |
| SA 1h | 6857.28 | 1240.65 | 1.36913 | 0.228215 |
| Me-JA 1h | 19887.5 | 2067.64 | 5.09686 | 0.294269 |
| Control 6h | 2824.29 | 555.082 | 0.575678 | 0.0881031 |
| ABA 6h | 9466.19 | 1754.43 | 1.81848 | 0.249972 |
| ACC 6h | 1909.89 | 110.666 | 0.351434 | 0.0652049 |
| BABA 6h | 1991.04 | 115.39 | 0.375245 | 0.0354808 |
| Chitin 6h | 2481.4 | 541.482 | 0.464873 | 0.141393 |
| Epi 6h | 2068.52 | 302.343 | 0.378974 | 0.0895758 |
| SA 6h | 1980.25 | 415.932 | 0.401304 | 0.0507016 |
| Me-JA 6h | 6516.39 | 1133.79 | 1.3376 | 0.286872 |
Source Transcript PGSC0003DMT400049666 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G15500.1 | +3 | 2e-94 | 293 | 132/151 (87%) | NAC domain containing protein 3 | chr3:5234731-5235882 FORWARD LENGTH=317 |
