Probe CUST_37634_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37634_PI426222305 | JHI_St_60k_v1 | DMT400049512 | TACATAGAAGGAAATGCACCGGGTGGTGATTTCACTCCTAGCCAATCTGCACAGATTACA |
All Microarray Probes Designed to Gene DMG401019239
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_37612_PI426222305 | JHI_St_60k_v1 | DMT400049515 | CCGAATGGGAGTCTTGATTCATTTATATTTGGTACTATTCTCCATCCAATATTGTGCAAA |
| CUST_37634_PI426222305 | JHI_St_60k_v1 | DMT400049512 | TACATAGAAGGAAATGCACCGGGTGGTGATTTCACTCCTAGCCAATCTGCACAGATTACA |
Microarray Signals from CUST_37634_PI426222305
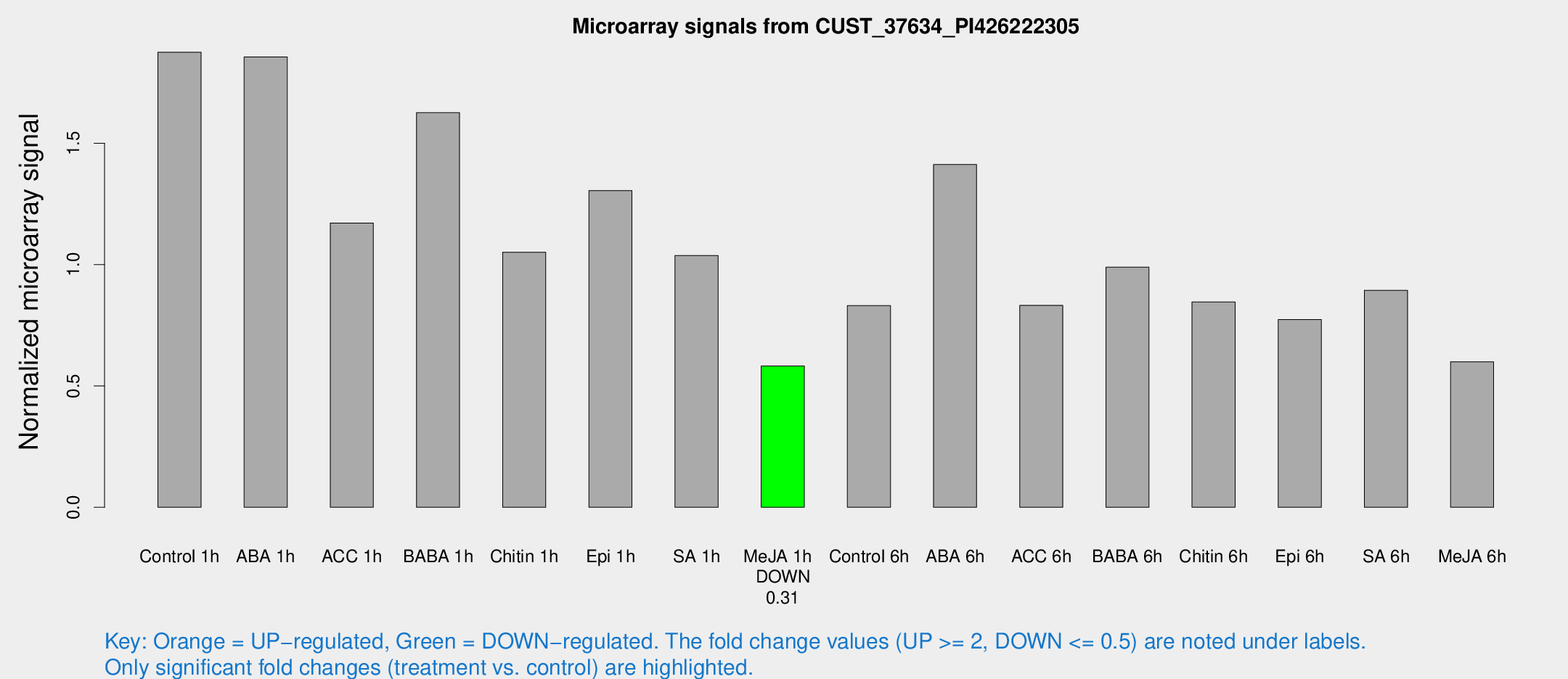
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 101.649 | 11.3238 | 1.87499 | 0.138286 |
| ABA 1h | 88.7644 | 8.31874 | 1.85559 | 0.131159 |
| ACC 1h | 72.2645 | 20.8932 | 1.17114 | 0.357888 |
| BABA 1h | 87.9245 | 17.834 | 1.62608 | 0.201601 |
| Chitin 1h | 50.8013 | 4.67886 | 1.05113 | 0.0964857 |
| Epi 1h | 61.7178 | 8.69487 | 1.30512 | 0.151339 |
| SA 1h | 57.4541 | 4.91925 | 1.03765 | 0.0891767 |
| Me-JA 1h | 25.7013 | 4.05102 | 0.582267 | 0.0934761 |
| Control 6h | 46.1665 | 7.86531 | 0.831041 | 0.0913382 |
| ABA 6h | 85.6408 | 19.6781 | 1.41234 | 0.332373 |
| ACC 6h | 51.6216 | 5.36891 | 0.832011 | 0.0848995 |
| BABA 6h | 63.9702 | 17.1573 | 0.989464 | 0.261098 |
| Chitin 6h | 49.294 | 6.95971 | 0.845927 | 0.12235 |
| Epi 6h | 51.042 | 13.3288 | 0.773949 | 0.205224 |
| SA 6h | 50.2152 | 12.2396 | 0.894223 | 0.149492 |
| Me-JA 6h | 33.2284 | 6.4892 | 0.599806 | 0.146872 |
Source Transcript PGSC0003DMT400049512 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G27290.1 | +1 | 0.0 | 818 | 405/736 (55%) | S-locus lectin protein kinase family protein | chr4:13666281-13669202 FORWARD LENGTH=783 |
