Probe CUST_36901_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36901_PI426222305 | JHI_St_60k_v1 | DMT400067455 | GATGTTTTACCTGATGAATGGAAGGCTGGCATTGCTTATGACACATACCTTATGCTAGAC |
All Microarray Probes Designed to Gene DMG400026220
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_36935_PI426222305 | JHI_St_60k_v1 | DMT400067456 | TATTATGGCAGCCCTGATTGCTATGGAACCCCTAGTGGAGCATGTGGATTTGGTGAATAT |
| CUST_36901_PI426222305 | JHI_St_60k_v1 | DMT400067455 | GATGTTTTACCTGATGAATGGAAGGCTGGCATTGCTTATGACACATACCTTATGCTAGAC |
Microarray Signals from CUST_36901_PI426222305
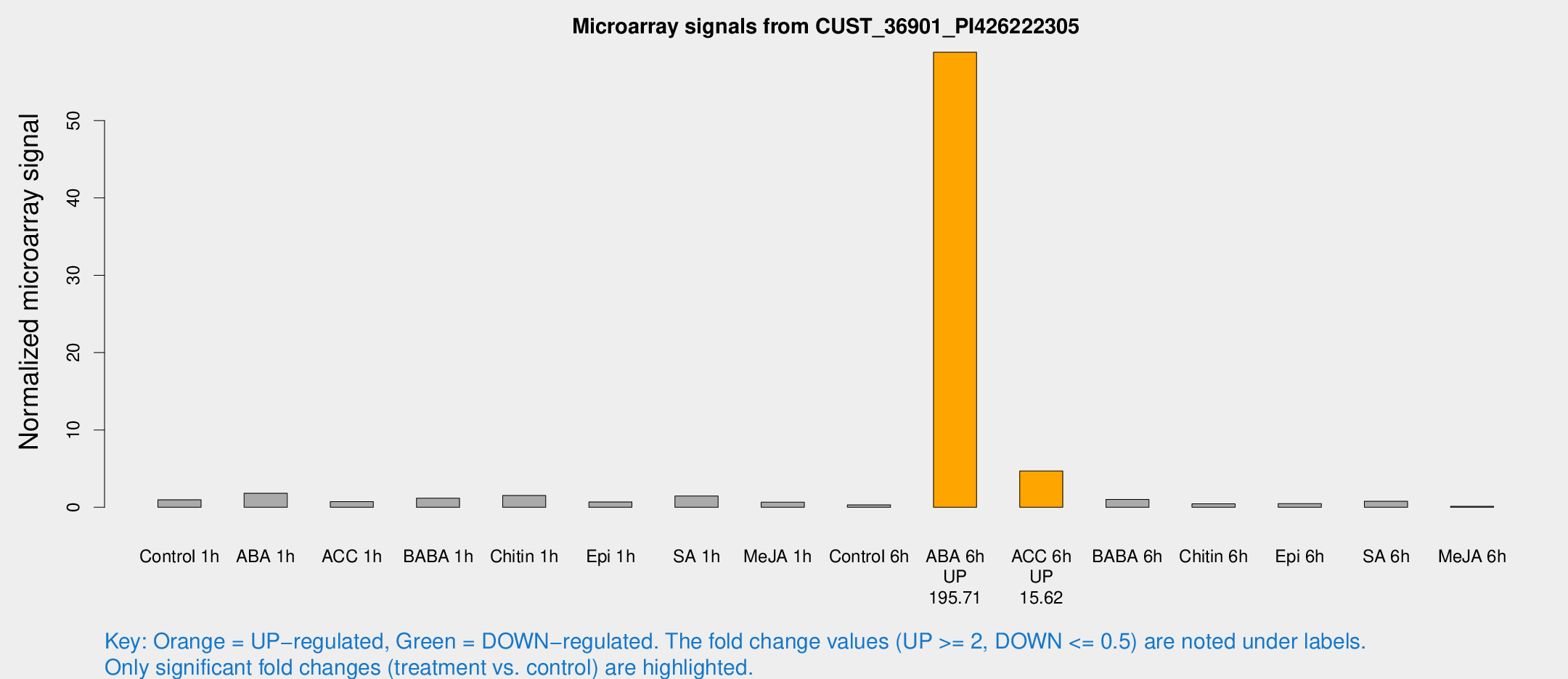
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 123.598 | 54.9697 | 0.976183 | 0.468924 |
| ABA 1h | 198.933 | 77.992 | 1.82284 | 1.18198 |
| ACC 1h | 92.7601 | 31.3991 | 0.742415 | 0.330555 |
| BABA 1h | 152.049 | 73.8819 | 1.1689 | 0.651472 |
| Chitin 1h | 147.448 | 36.6215 | 1.5288 | 0.405548 |
| Epi 1h | 67.9235 | 23.3371 | 0.694746 | 0.257499 |
| SA 1h | 217.639 | 91.4795 | 1.46791 | 1.53033 |
| Me-JA 1h | 60.758 | 17.8472 | 0.662223 | 0.201616 |
| Control 6h | 35.7784 | 13.2171 | 0.300589 | 0.110321 |
| ABA 6h | 6585.06 | 1205.69 | 58.8297 | 8.82432 |
| ACC 6h | 557.723 | 83.2838 | 4.6958 | 0.273845 |
| BABA 6h | 161.645 | 94.6947 | 1.01584 | 0.697129 |
| Chitin 6h | 82.1893 | 37.192 | 0.44567 | 0.926234 |
| Epi 6h | 56.0174 | 6.76974 | 0.482463 | 0.0479993 |
| SA 6h | 92.5636 | 40.097 | 0.773074 | 0.283038 |
| Me-JA 6h | 15.7635 | 7.09095 | 0.126864 | 0.0611633 |
Source Transcript PGSC0003DMT400067455 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G17030.1 | +1 | 2e-82 | 248 | 116/205 (57%) | expansin-like B1 | chr4:9581817-9583181 REVERSE LENGTH=250 |
