Probe CUST_3535_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3535_PI426222305 | JHI_St_60k_v1 | DMT400064256 | ATCAAAGGATCACCACATCCAAGGGGATATTACAAGTGTAGTAGTGTAAGAGGTTGTCCA |
All Microarray Probes Designed to Gene DMG400024961
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3532_PI426222305 | JHI_St_60k_v1 | DMT400064255 | GCCAATCAAAGGATCACCACATCCAAGGTATATTTAGAGCTAATTGATCATGAATTTGTT |
| CUST_3535_PI426222305 | JHI_St_60k_v1 | DMT400064256 | ATCAAAGGATCACCACATCCAAGGGGATATTACAAGTGTAGTAGTGTAAGAGGTTGTCCA |
Microarray Signals from CUST_3535_PI426222305
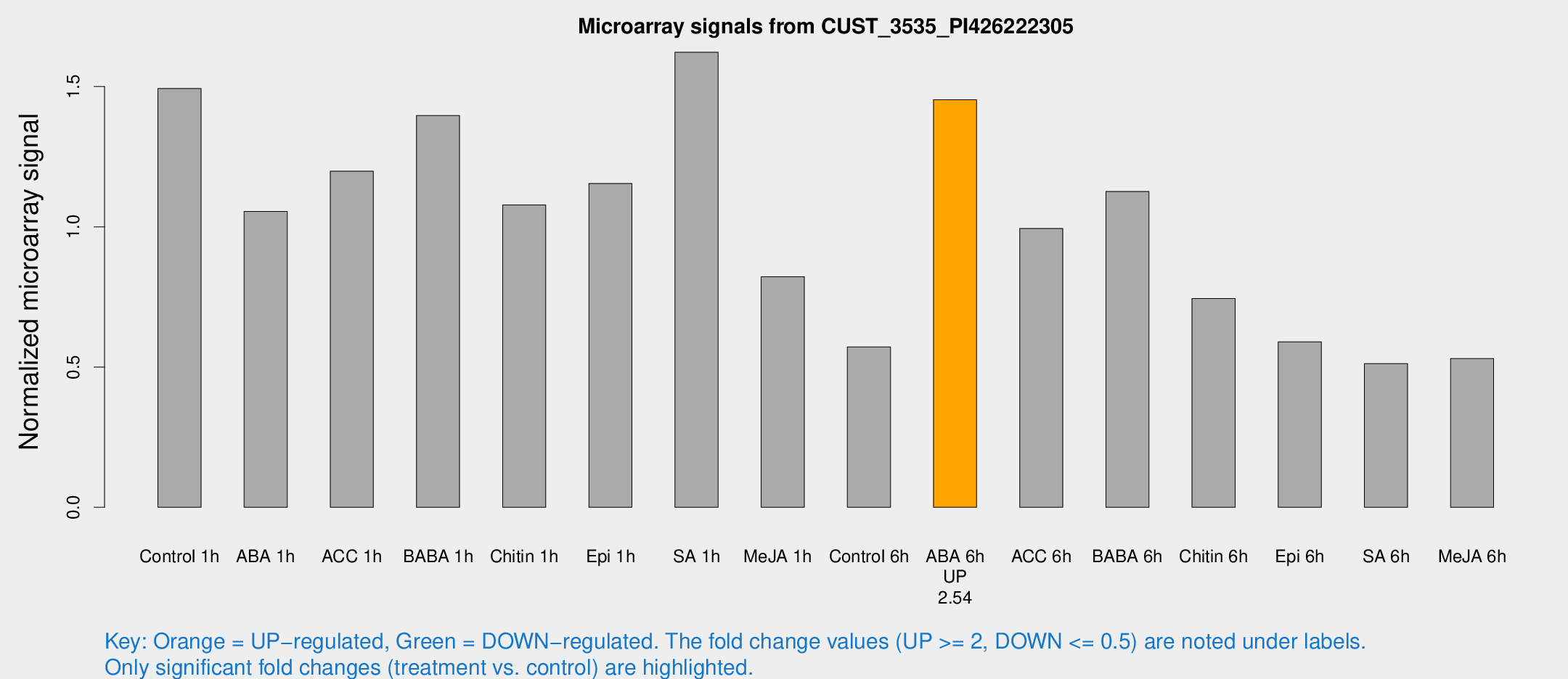
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2409.97 | 405.123 | 1.49288 | 0.155735 |
| ABA 1h | 1503.72 | 235.554 | 1.05507 | 0.0899733 |
| ACC 1h | 2353.99 | 821.639 | 1.19834 | 0.56445 |
| BABA 1h | 2155.08 | 274.674 | 1.39652 | 0.0806675 |
| Chitin 1h | 1534.38 | 125.868 | 1.07799 | 0.0622877 |
| Epi 1h | 1584.14 | 143.042 | 1.15455 | 0.0726369 |
| SA 1h | 2631.66 | 163.227 | 1.62246 | 0.0936968 |
| Me-JA 1h | 1070.78 | 131.845 | 0.822132 | 0.0578052 |
| Control 6h | 1004.78 | 280.85 | 0.571856 | 0.156535 |
| ABA 6h | 2524.85 | 480.374 | 1.45276 | 0.214333 |
| ACC 6h | 1808.42 | 161.805 | 0.994111 | 0.0983539 |
| BABA 6h | 2036.67 | 347.051 | 1.12622 | 0.149317 |
| Chitin 6h | 1249.54 | 72.422 | 0.744596 | 0.0507753 |
| Epi 6h | 1053.56 | 86.4599 | 0.590169 | 0.0341535 |
| SA 6h | 811.525 | 104.744 | 0.512058 | 0.0296764 |
| Me-JA 6h | 870.412 | 196.884 | 0.530387 | 0.0850368 |
Source Transcript PGSC0003DMT400064256 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G23320.1 | +2 | 2e-66 | 218 | 159/335 (47%) | WRKY DNA-binding protein 15 | chr2:9924998-9926154 FORWARD LENGTH=317 |
