Probe CUST_34231_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34231_PI426222305 | JHI_St_60k_v1 | DMT400044818 | CTTGTTGTGTTACATCCTGGGCTATATTTTGCTAATGACTAATAATATACCCTACTCTTC |
All Microarray Probes Designed to Gene DMG400017394
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34248_PI426222305 | JHI_St_60k_v1 | DMT400044819 | CTTGTTGTGTTACATCCTGGGCTATATTTTGCTAATGACTAATAATATACCCTACTCTTC |
| CUST_34231_PI426222305 | JHI_St_60k_v1 | DMT400044818 | CTTGTTGTGTTACATCCTGGGCTATATTTTGCTAATGACTAATAATATACCCTACTCTTC |
Microarray Signals from CUST_34231_PI426222305
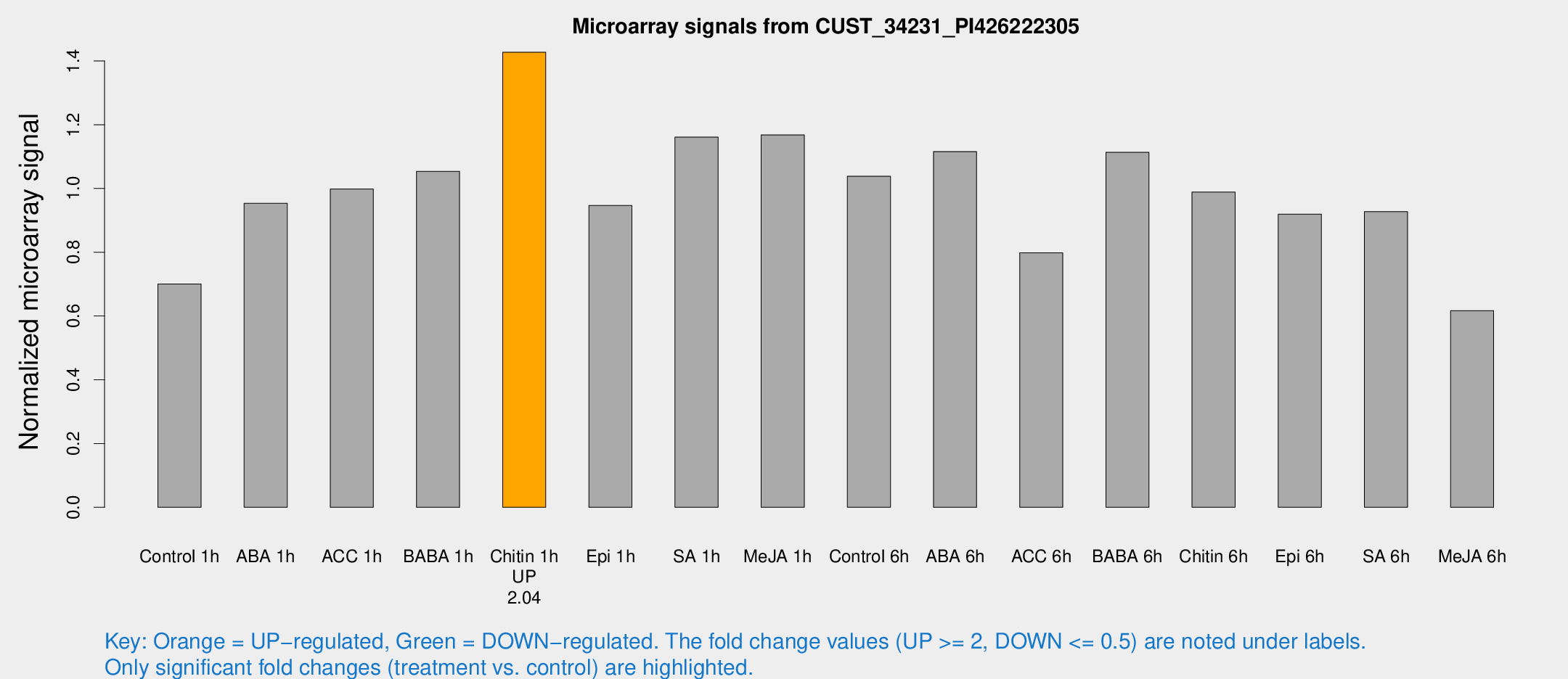
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3692.19 | 240.156 | 0.700522 | 0.0404499 |
| ABA 1h | 4518.72 | 664.096 | 0.953505 | 0.0709275 |
| ACC 1h | 5407.78 | 381.426 | 0.998176 | 0.135604 |
| BABA 1h | 5560.25 | 1052.86 | 1.05378 | 0.115214 |
| Chitin 1h | 6948.54 | 1247.88 | 1.42731 | 0.240262 |
| Epi 1h | 4317.16 | 271.481 | 0.946648 | 0.0546601 |
| SA 1h | 6408.86 | 1026.28 | 1.16086 | 0.116936 |
| Me-JA 1h | 5024.53 | 322.622 | 1.16771 | 0.0674231 |
| Control 6h | 5764.84 | 1207.54 | 1.03851 | 0.158871 |
| ABA 6h | 6288.93 | 648.919 | 1.11587 | 0.0644285 |
| ACC 6h | 4980.25 | 903.164 | 0.79798 | 0.0860719 |
| BABA 6h | 6593.3 | 579.089 | 1.11342 | 0.070957 |
| Chitin 6h | 5541.03 | 321.004 | 0.988594 | 0.0570811 |
| Epi 6h | 5461.45 | 342.149 | 0.919478 | 0.0881432 |
| SA 6h | 4944.79 | 756.565 | 0.92737 | 0.0535477 |
| Me-JA 6h | 3533.31 | 949.727 | 0.617003 | 0.152168 |
Source Transcript PGSC0003DMT400044818 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G35790.1 | +1 | 0.0 | 871 | 429/529 (81%) | glucose-6-phosphate dehydrogenase 1 | chr5:13956879-13959686 REVERSE LENGTH=576 |
