Probe CUST_34186_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34186_PI426222305 | JHI_St_60k_v1 | DMT400041150 | TTTCTCTCGCAATTTATCTCATGGATTCTCAACACAAGAGTAATCGTCAACTCCAAAGAG |
All Microarray Probes Designed to Gene DMG400015925
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_34186_PI426222305 | JHI_St_60k_v1 | DMT400041150 | TTTCTCTCGCAATTTATCTCATGGATTCTCAACACAAGAGTAATCGTCAACTCCAAAGAG |
| CUST_34204_PI426222305 | JHI_St_60k_v1 | DMT400041149 | TGCTAAACACCCTTCAATTTTAAGAGAATCGATTAGTCCGTATCCATTCCAGTGTAGTCA |
Microarray Signals from CUST_34186_PI426222305
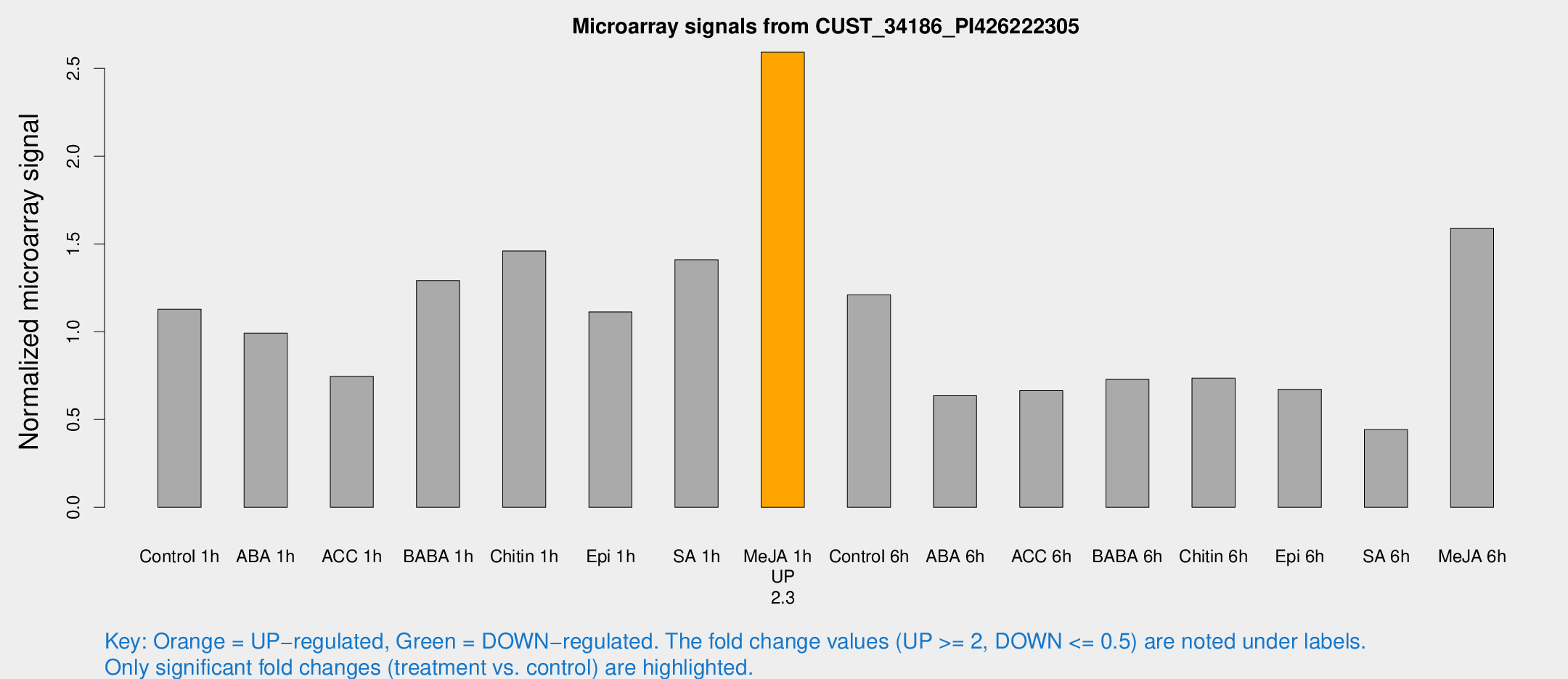
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 196.791 | 20.8957 | 1.12858 | 0.0677409 |
| ABA 1h | 152.416 | 12.7558 | 0.991646 | 0.0605929 |
| ACC 1h | 143.793 | 43.971 | 0.74599 | 0.202699 |
| BABA 1h | 219.769 | 33.8727 | 1.2914 | 0.120535 |
| Chitin 1h | 229.479 | 25.1824 | 1.45991 | 0.282147 |
| Epi 1h | 168.17 | 18.8205 | 1.11282 | 0.128323 |
| SA 1h | 251.433 | 19.4102 | 1.40978 | 0.199929 |
| Me-JA 1h | 367.884 | 32.9401 | 2.59179 | 0.171108 |
| Control 6h | 219.955 | 44.8902 | 1.20981 | 0.175958 |
| ABA 6h | 123.282 | 26.0969 | 0.63601 | 0.134452 |
| ACC 6h | 137.349 | 28.5667 | 0.664131 | 0.0889296 |
| BABA 6h | 146.486 | 27.8179 | 0.728984 | 0.129297 |
| Chitin 6h | 138.138 | 19.7819 | 0.735344 | 0.111497 |
| Epi 6h | 152.585 | 55.3959 | 0.671864 | 0.35815 |
| SA 6h | 92.6942 | 36.8923 | 0.442337 | 0.287458 |
| Me-JA 6h | 310.869 | 93.4514 | 1.58984 | 0.531623 |
Source Transcript PGSC0003DMT400041150 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G43020.1 | +1 | 0.0 | 794 | 390/490 (80%) | polyamine oxidase 2 | chr2:17891945-17894440 FORWARD LENGTH=490 |
