Probe CUST_33300_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33300_PI426222305 | JHI_St_60k_v1 | DMT400017914 | GTTGTTGTAATAATGACTCTGTTCTGCAGAGGGAATTGGAATAAATAATATTGGAGCAAC |
All Microarray Probes Designed to Gene DMG400006956
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_33248_PI426222305 | JHI_St_60k_v1 | DMT400017915 | GTTGTTGTAATAATGACTCTGTTCTGCAGAGGGAATTGGAATAAATAATATTGGAGCAAC |
| CUST_33300_PI426222305 | JHI_St_60k_v1 | DMT400017914 | GTTGTTGTAATAATGACTCTGTTCTGCAGAGGGAATTGGAATAAATAATATTGGAGCAAC |
Microarray Signals from CUST_33300_PI426222305
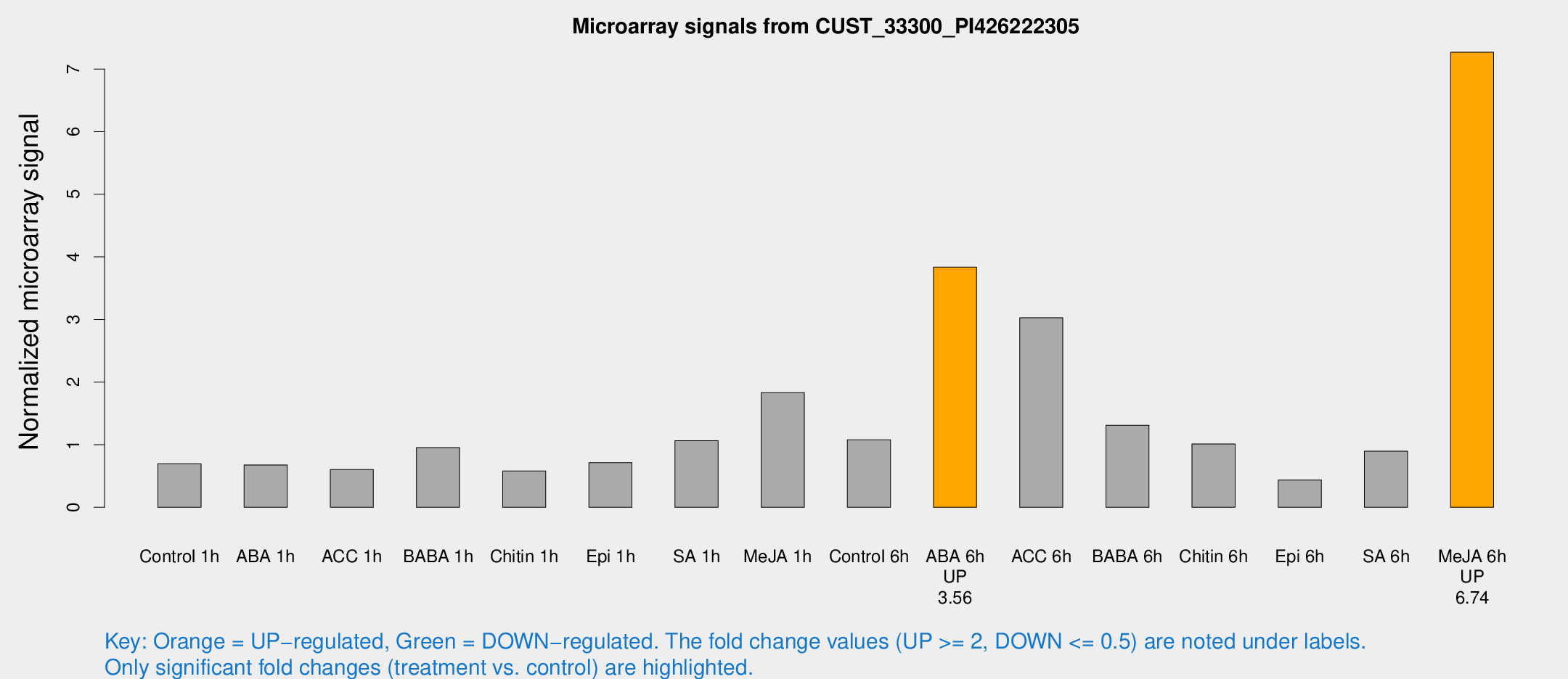
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5470.37 | 1031.92 | 0.695271 | 0.166964 |
| ABA 1h | 4913.45 | 1213.72 | 0.677177 | 0.173515 |
| ACC 1h | 6608.84 | 2869.22 | 0.603246 | 0.476927 |
| BABA 1h | 7154.2 | 1102.99 | 0.953115 | 0.077189 |
| Chitin 1h | 4309.24 | 1168.82 | 0.580094 | 0.171629 |
| Epi 1h | 5180.39 | 1588.94 | 0.712072 | 0.24562 |
| SA 1h | 9477.7 | 3526.19 | 1.06465 | 0.447249 |
| Me-JA 1h | 11639.8 | 1786.47 | 1.83114 | 0.12937 |
| Control 6h | 9664.05 | 4084.94 | 1.07758 | 0.384948 |
| ABA 6h | 31767.9 | 4760.82 | 3.83464 | 0.620678 |
| ACC 6h | 28961.4 | 8856.99 | 3.02815 | 1.21929 |
| BABA 6h | 11692 | 2587.73 | 1.31135 | 0.26785 |
| Chitin 6h | 9764.79 | 3989.64 | 1.01106 | 0.523842 |
| Epi 6h | 4089.73 | 1114.96 | 0.435085 | 0.19269 |
| SA 6h | 6997.14 | 1393.76 | 0.896933 | 0.128277 |
| Me-JA 6h | 55743.5 | 6175.66 | 7.26824 | 1.08882 |
Source Transcript PGSC0003DMT400017914 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G01500.2 | +3 | 6e-84 | 265 | 124/176 (70%) | carbonic anhydrase 1 | chr3:194853-197873 REVERSE LENGTH=347 |
