Probe CUST_32865_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32865_PI426222305 | JHI_St_60k_v1 | DMT400062618 | GGATATCAATTCCATCATTTCTCACCATGTAAGCTTAATCCCCAATTCAATTCTCTCTGT |
All Microarray Probes Designed to Gene DMG400024368
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32882_PI426222305 | JHI_St_60k_v1 | DMT400062617 | CCAAAGATTCAGAAATCATATACACATGGCTAGTGTTGAAGCAAGACATCTCAATCTTTT |
| CUST_32865_PI426222305 | JHI_St_60k_v1 | DMT400062618 | GGATATCAATTCCATCATTTCTCACCATGTAAGCTTAATCCCCAATTCAATTCTCTCTGT |
Microarray Signals from CUST_32865_PI426222305
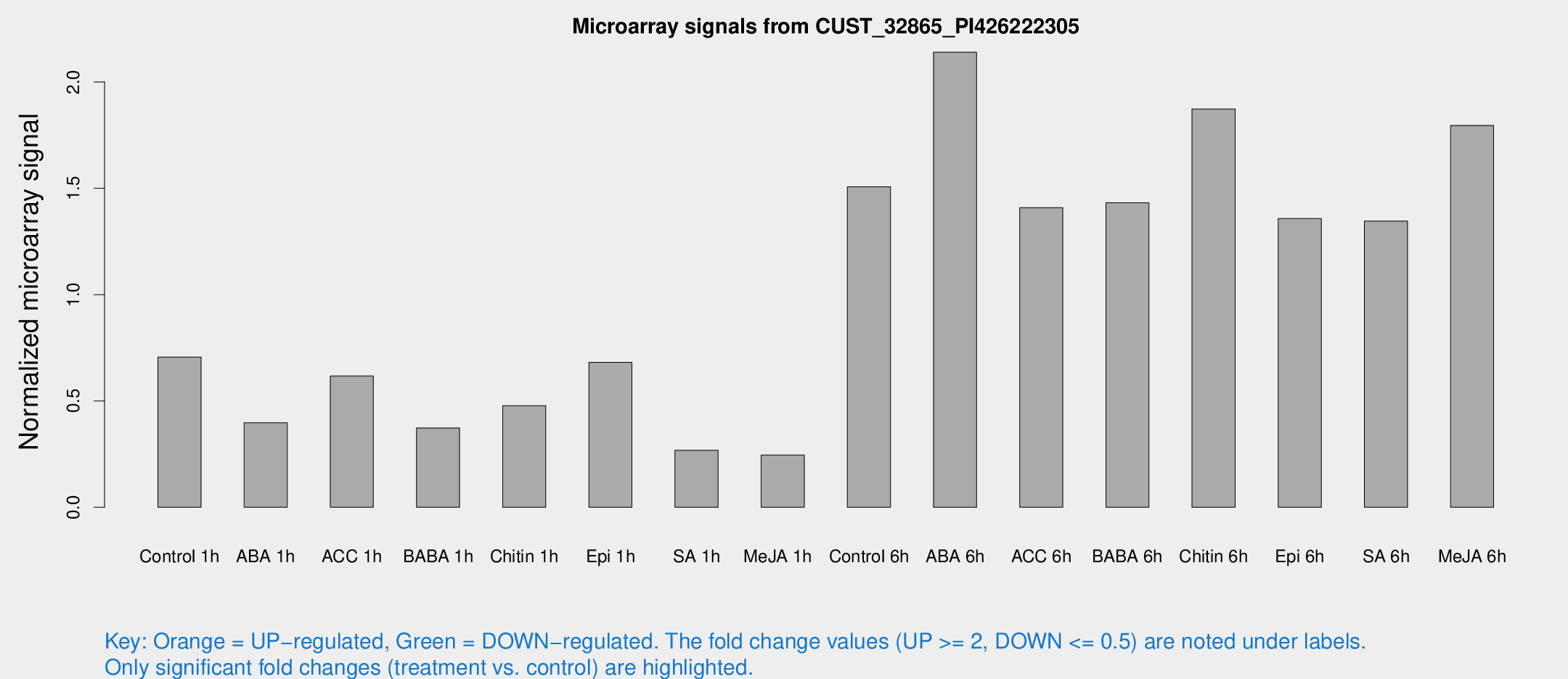
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 39.4774 | 8.43045 | 0.706613 | 0.108714 |
| ABA 1h | 20.8849 | 5.88178 | 0.397547 | 0.125386 |
| ACC 1h | 47.6863 | 22.8289 | 0.617317 | 0.462412 |
| BABA 1h | 21.7224 | 6.49054 | 0.372526 | 0.106307 |
| Chitin 1h | 25.0624 | 7.52891 | 0.477438 | 0.120961 |
| Epi 1h | 31.6469 | 3.44406 | 0.681207 | 0.0746834 |
| SA 1h | 19.7196 | 9.58936 | 0.267981 | 0.186274 |
| Me-JA 1h | 11.4773 | 3.0811 | 0.245841 | 0.0734445 |
| Control 6h | 88.4177 | 24.6862 | 1.50664 | 0.360046 |
| ABA 6h | 124.046 | 15.6996 | 2.13963 | 0.19145 |
| ACC 6h | 92.786 | 21.8476 | 1.40932 | 0.35349 |
| BABA 6h | 88.5865 | 16.1249 | 1.43221 | 0.23541 |
| Chitin 6h | 107.384 | 8.11833 | 1.87259 | 0.123383 |
| Epi 6h | 82.2663 | 5.91454 | 1.35748 | 0.0970309 |
| SA 6h | 71.5251 | 5.25057 | 1.34577 | 0.234389 |
| Me-JA 6h | 104.504 | 32.3744 | 1.79535 | 0.401963 |
Source Transcript PGSC0003DMT400062618 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G12920.1 | +2 | 3e-71 | 229 | 143/347 (41%) | SBP (S-ribonuclease binding protein) family protein | chr3:4122127-4123323 REVERSE LENGTH=335 |
