Probe CUST_32654_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32654_PI426222305 | JHI_St_60k_v1 | DMT400047278 | GTAATTTCAGCTTGGAAGTTGATTAAGGAGTCAGCCATTGCTTTGCTGAGAAAAAGTGTG |
All Microarray Probes Designed to Gene DMG400018358
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_32654_PI426222305 | JHI_St_60k_v1 | DMT400047278 | GTAATTTCAGCTTGGAAGTTGATTAAGGAGTCAGCCATTGCTTTGCTGAGAAAAAGTGTG |
Microarray Signals from CUST_32654_PI426222305
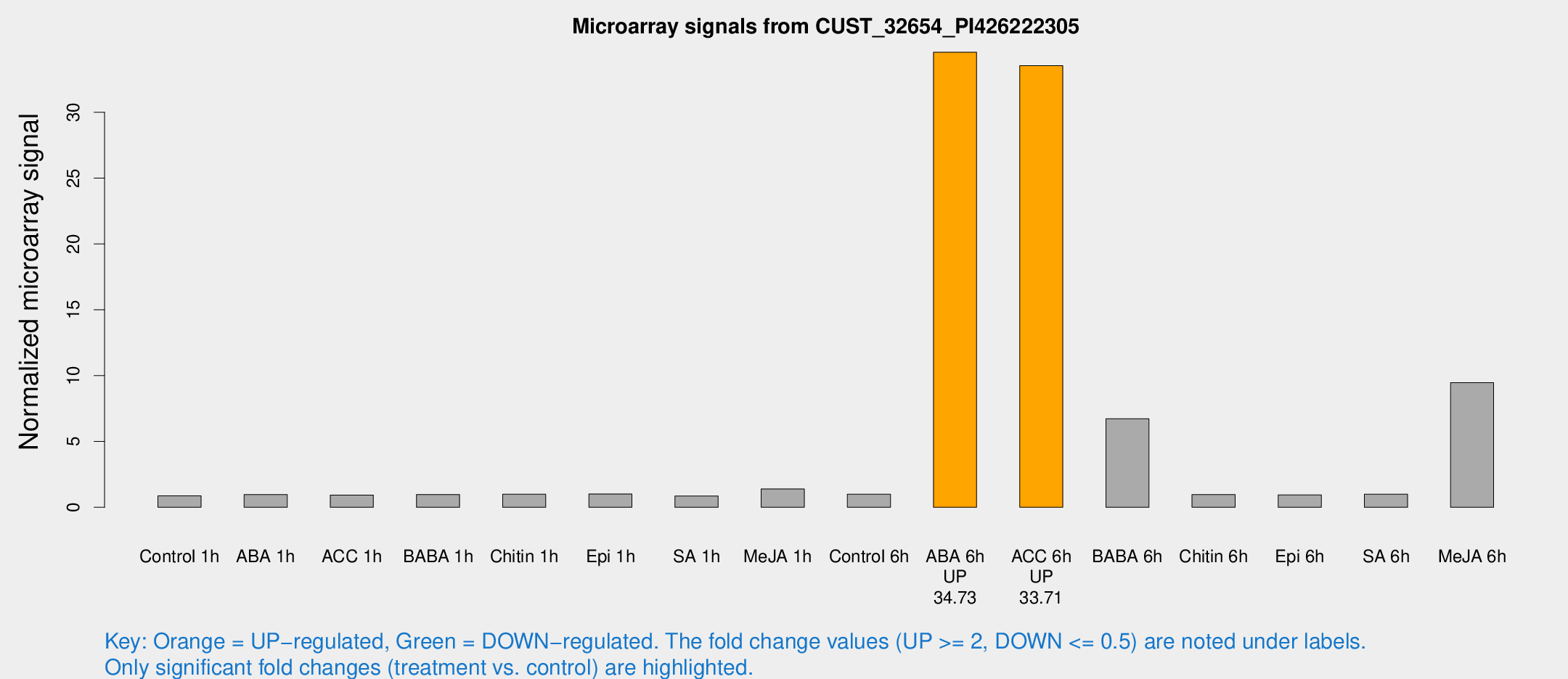
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.81715 | 3.36943 | 0.865504 | 0.501172 |
| ABA 1h | 5.73024 | 3.32839 | 0.962734 | 0.558419 |
| ACC 1h | 6.43941 | 3.74051 | 0.931436 | 0.540139 |
| BABA 1h | 6.3082 | 3.6545 | 0.973547 | 0.56374 |
| Chitin 1h | 6.01577 | 3.49501 | 0.992517 | 0.574863 |
| Epi 1h | 5.90629 | 3.42645 | 1.01375 | 0.587032 |
| SA 1h | 5.90476 | 3.42606 | 0.855049 | 0.495945 |
| Me-JA 1h | 7.80236 | 3.55427 | 1.39232 | 0.654629 |
| Control 6h | 6.71611 | 3.68053 | 0.995074 | 0.547791 |
| ABA 6h | 321.286 | 125.703 | 34.5624 | 23.5945 |
| ACC 6h | 297.984 | 118.017 | 33.5463 | 18.1388 |
| BABA 6h | 67.3179 | 31.631 | 6.72799 | 4.92696 |
| Chitin 6h | 6.87884 | 3.98372 | 0.961705 | 0.556938 |
| Epi 6h | 7.18799 | 4.20365 | 0.94056 | 0.54545 |
| SA 6h | 6.61298 | 3.8366 | 0.994645 | 0.576823 |
| Me-JA 6h | 146.324 | 85.4566 | 9.47172 | 46.8241 |
Source Transcript PGSC0003DMT400047278 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G20340.1 | +3 | 0.0 | 555 | 263/481 (55%) | Pyridoxal phosphate (PLP)-dependent transferases superfamily protein | chr2:8779804-8782490 FORWARD LENGTH=490 |
