Probe CUST_30339_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30339_PI426222305 | JHI_St_60k_v1 | DMT400069837 | GCCATCTTATGTTGTGTTGGTAAGCTGATGTTGAATGTTGAAAAAGGAGTAATAGATACT |
All Microarray Probes Designed to Gene DMG400027154
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30347_PI426222305 | JHI_St_60k_v1 | DMT400069836 | AGGAGAAGTAGCTGTCCGATTTTTTATTGGTCTTGTTAAAATCCTACCTGCCAAATATGT |
| CUST_30339_PI426222305 | JHI_St_60k_v1 | DMT400069837 | GCCATCTTATGTTGTGTTGGTAAGCTGATGTTGAATGTTGAAAAAGGAGTAATAGATACT |
Microarray Signals from CUST_30339_PI426222305
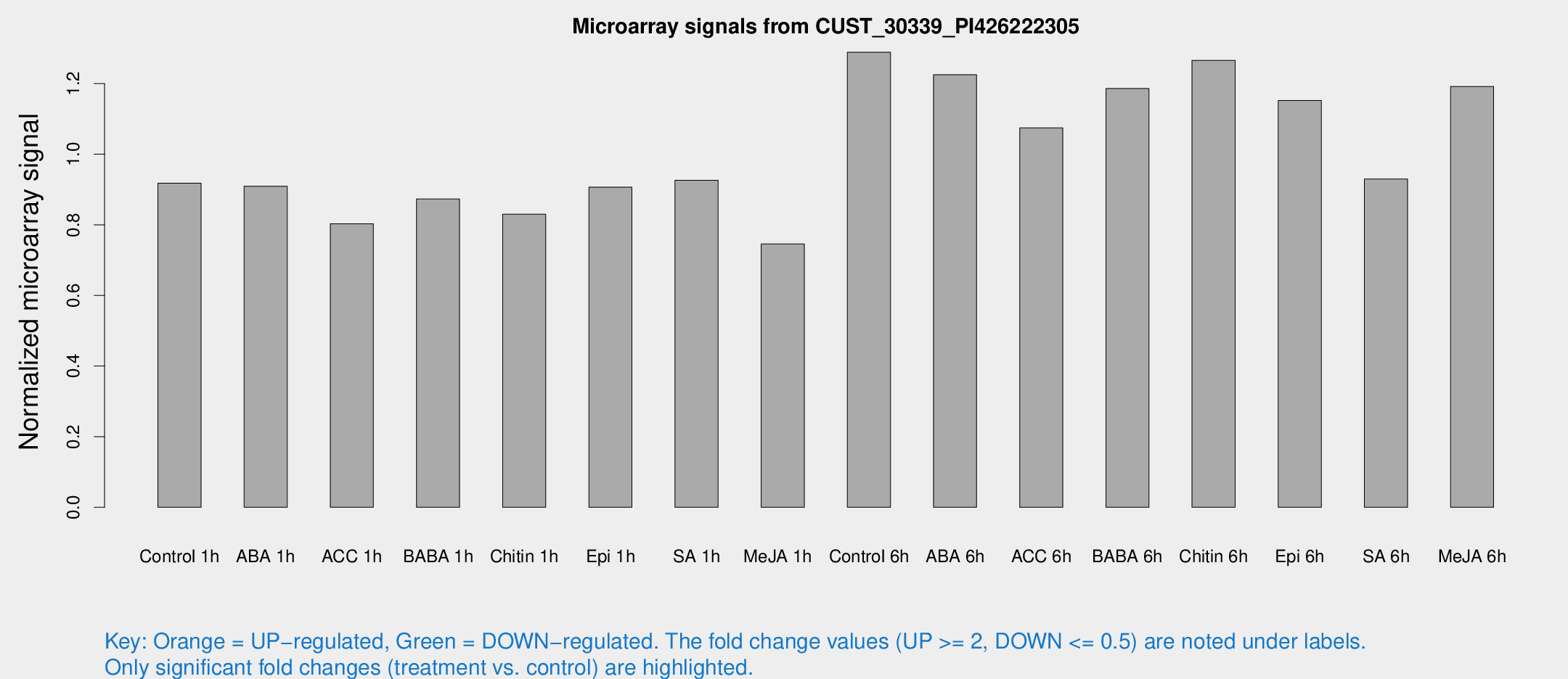
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 502.396 | 91.8104 | 0.917974 | 0.121067 |
| ABA 1h | 426.186 | 26.4746 | 0.909311 | 0.0528762 |
| ACC 1h | 454.02 | 86.3675 | 0.802822 | 0.113025 |
| BABA 1h | 460.875 | 82.3654 | 0.872994 | 0.0832667 |
| Chitin 1h | 394.282 | 22.9949 | 0.829873 | 0.0500683 |
| Epi 1h | 419.082 | 44.309 | 0.906477 | 0.0918697 |
| SA 1h | 505.319 | 40.2876 | 0.925887 | 0.0537547 |
| Me-JA 1h | 323.273 | 25.9035 | 0.745599 | 0.0436375 |
| Control 6h | 727.5 | 165.03 | 1.28866 | 0.230777 |
| ABA 6h | 692.699 | 61.448 | 1.22496 | 0.0709612 |
| ACC 6h | 661.071 | 85.7269 | 1.07445 | 0.0623221 |
| BABA 6h | 706.633 | 64.527 | 1.18592 | 0.0800161 |
| Chitin 6h | 713.892 | 44.6307 | 1.26578 | 0.0733384 |
| Epi 6h | 689.094 | 50.9235 | 1.15202 | 0.123549 |
| SA 6h | 523.673 | 127.588 | 0.92964 | 0.164869 |
| Me-JA 6h | 649.284 | 115.208 | 1.1918 | 0.136531 |
Source Transcript PGSC0003DMT400069837 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G06440.1 | +2 | 0.0 | 585 | 325/650 (50%) | Galactosyltransferase family protein | chr3:1972913-1975272 REVERSE LENGTH=619 |
