Probe CUST_3027_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3027_PI426222305 | JHI_St_60k_v1 | DMT400000652 | GTAGTAGCAAATTTTCCGTGACTGTCTATAGTAGTACACATTTTCTATCATAATCAAGGC |
All Microarray Probes Designed to Gene DMG400000232
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_3027_PI426222305 | JHI_St_60k_v1 | DMT400000652 | GTAGTAGCAAATTTTCCGTGACTGTCTATAGTAGTACACATTTTCTATCATAATCAAGGC |
| CUST_3221_PI426222305 | JHI_St_60k_v1 | DMT400000653 | GTAGTAGCAAATTTTCCGTGACTGTCTATAGTAGTACACATTTTCTATCATAATCAAGGC |
Microarray Signals from CUST_3027_PI426222305
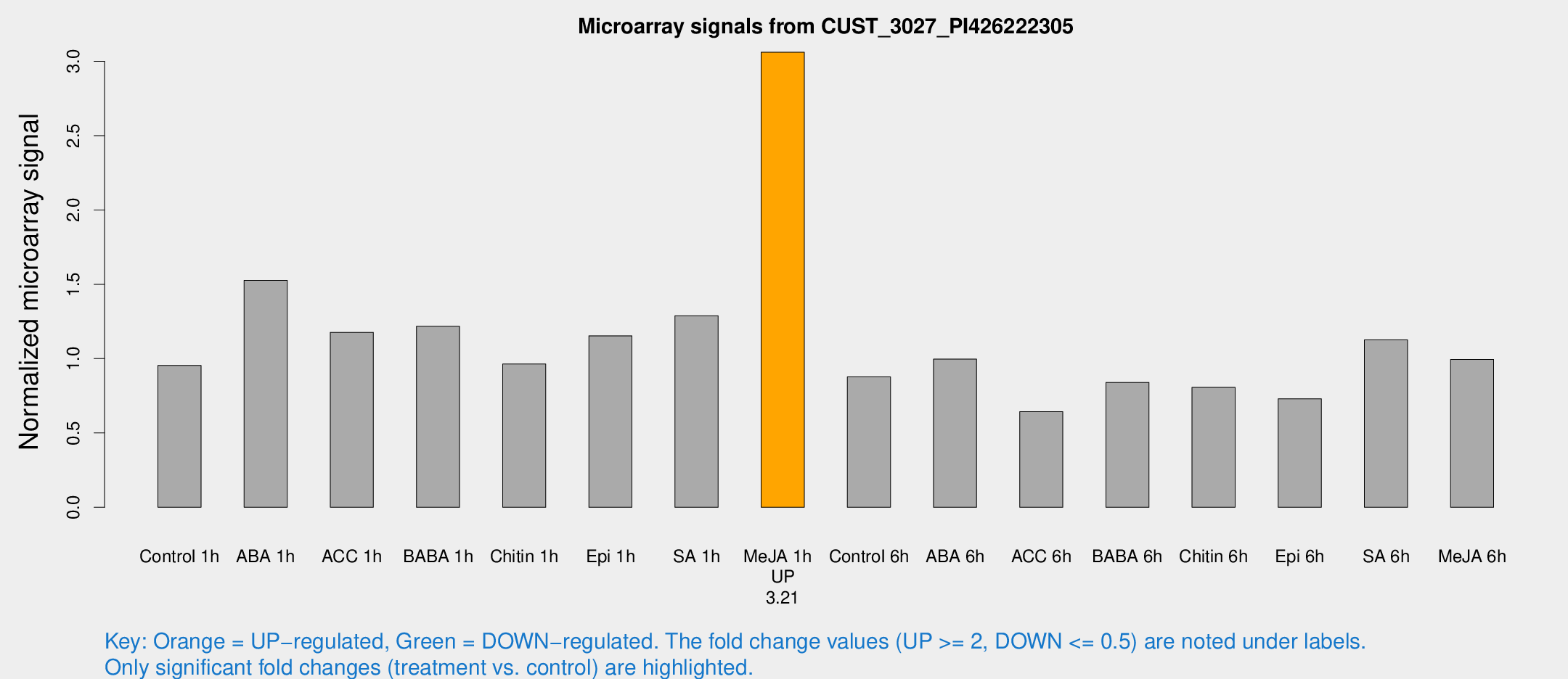
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 668.66 | 137.834 | 0.954457 | 0.184459 |
| ABA 1h | 980.214 | 278.642 | 1.52642 | 0.33286 |
| ACC 1h | 878.48 | 251.993 | 1.17675 | 0.28375 |
| BABA 1h | 796.805 | 87.4883 | 1.21732 | 0.0705002 |
| Chitin 1h | 586.509 | 58.7956 | 0.963715 | 0.0558933 |
| Epi 1h | 693.93 | 121.852 | 1.15351 | 0.21247 |
| SA 1h | 887.237 | 51.3298 | 1.28813 | 0.0745128 |
| Me-JA 1h | 1696.97 | 195.735 | 3.06077 | 0.176815 |
| Control 6h | 599.315 | 74.5172 | 0.877492 | 0.0557294 |
| ABA 6h | 732.249 | 133.138 | 0.996399 | 0.120849 |
| ACC 6h | 501.091 | 53.5847 | 0.643592 | 0.0573705 |
| BABA 6h | 638.74 | 71.5075 | 0.840029 | 0.0635726 |
| Chitin 6h | 575.886 | 33.4863 | 0.806313 | 0.0487001 |
| Epi 6h | 552.503 | 32.1994 | 0.729323 | 0.0908642 |
| SA 6h | 749.787 | 50.0351 | 1.12563 | 0.155711 |
| Me-JA 6h | 671.821 | 70.6929 | 0.99444 | 0.0576382 |
Source Transcript PGSC0003DMT400000652 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G01260.1 | +2 | 0.0 | 538 | 315/627 (50%) | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | chr1:109595-111367 FORWARD LENGTH=590 |
