Probe CUST_30228_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30228_PI426222305 | JHI_St_60k_v1 | DMT400012159 | AAACTCTTGCGTGAAAGCTGGACCATCAGTTAGAGGAGCGGCATGGGGAAAACTTAATTT |
All Microarray Probes Designed to Gene DMG400004766
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_30228_PI426222305 | JHI_St_60k_v1 | DMT400012159 | AAACTCTTGCGTGAAAGCTGGACCATCAGTTAGAGGAGCGGCATGGGGAAAACTTAATTT |
| CUST_30258_PI426222305 | JHI_St_60k_v1 | DMT400012160 | TGGTTGTTTTGATGAGAAACTCTTGCGCCAGAGAGAAAGCCCAAAATTTTCTAGTGTAAT |
Microarray Signals from CUST_30228_PI426222305
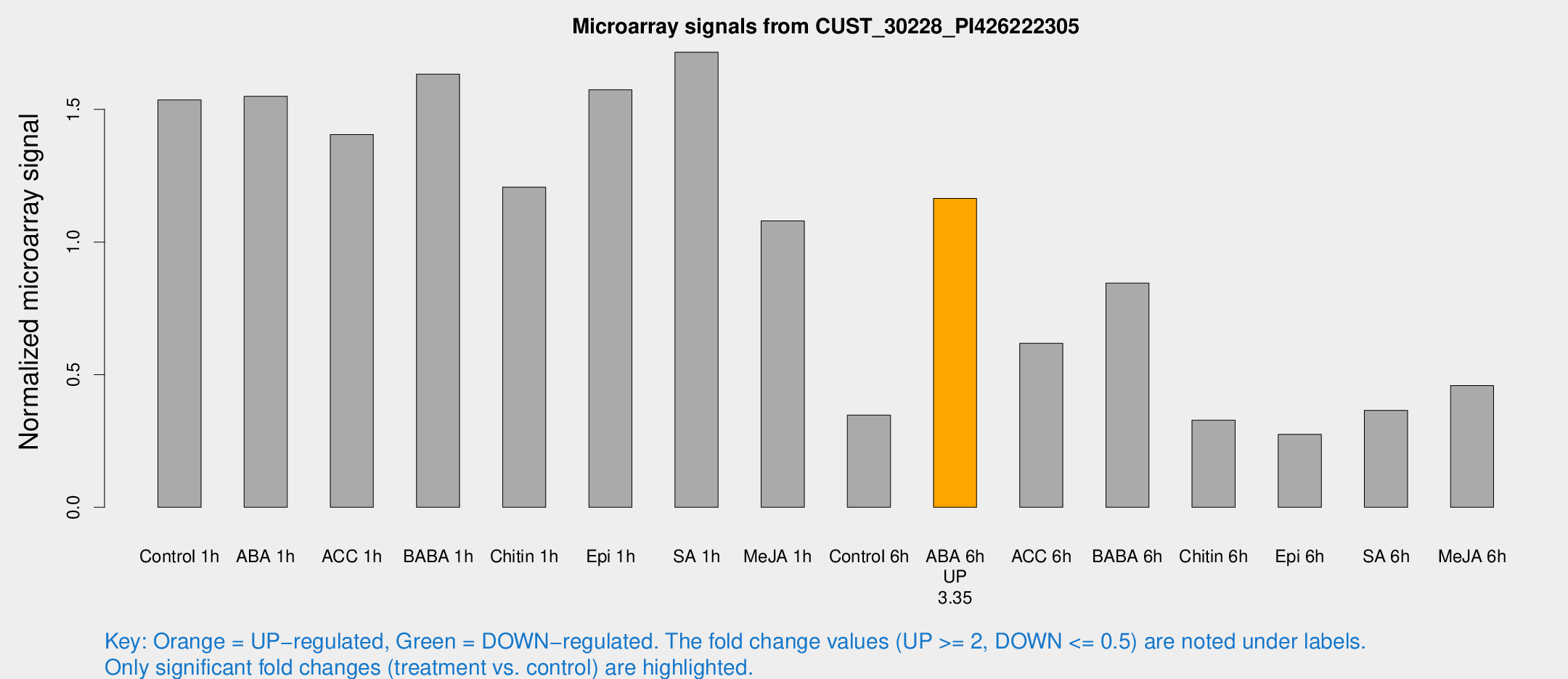
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 824.993 | 144.086 | 1.53582 | 0.179456 |
| ABA 1h | 723.342 | 71.9486 | 1.54932 | 0.0897156 |
| ACC 1h | 822.351 | 211.276 | 1.40518 | 0.346945 |
| BABA 1h | 875.22 | 203.365 | 1.63294 | 0.26542 |
| Chitin 1h | 595.537 | 137.066 | 1.20693 | 0.17541 |
| Epi 1h | 715.233 | 51.2668 | 1.5738 | 0.0950656 |
| SA 1h | 1015.25 | 272.709 | 1.71561 | 0.513088 |
| Me-JA 1h | 469.209 | 61.7967 | 1.07992 | 0.0678681 |
| Control 6h | 205.841 | 61.4306 | 0.347694 | 0.106123 |
| ABA 6h | 690.486 | 166.221 | 1.1648 | 0.252693 |
| ACC 6h | 370.827 | 21.8088 | 0.618456 | 0.0719606 |
| BABA 6h | 505.221 | 78.3296 | 0.845055 | 0.10671 |
| Chitin 6h | 183.084 | 11.2464 | 0.32837 | 0.020134 |
| Epi 6h | 178.174 | 49.7672 | 0.274991 | 0.0783259 |
| SA 6h | 192.734 | 26.5045 | 0.365361 | 0.0223019 |
| Me-JA 6h | 240.648 | 21.3062 | 0.459283 | 0.0273044 |
Source Transcript PGSC0003DMT400012159 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G13200.1 | +2 | 2e-112 | 333 | 147/231 (64%) | GRAM domain family protein | chr5:4207081-4208079 FORWARD LENGTH=272 |
