Probe CUST_29827_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29827_PI426222305 | JHI_St_60k_v1 | DMT400033409 | GCCGCTGGTATACCTAGAGTTTGTGGTCTAGTGCTATTAAAGGCATTAATGTGGGCAAAG |
All Microarray Probes Designed to Gene DMG400012837
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29833_PI426222305 | JHI_St_60k_v1 | DMT400033408 | ATTCCTTACAAGATCAGCCCCTCCACTGACTGCTCCAAGTACGTTATTTATATTTTGTGA |
| CUST_29827_PI426222305 | JHI_St_60k_v1 | DMT400033409 | GCCGCTGGTATACCTAGAGTTTGTGGTCTAGTGCTATTAAAGGCATTAATGTGGGCAAAG |
Microarray Signals from CUST_29827_PI426222305
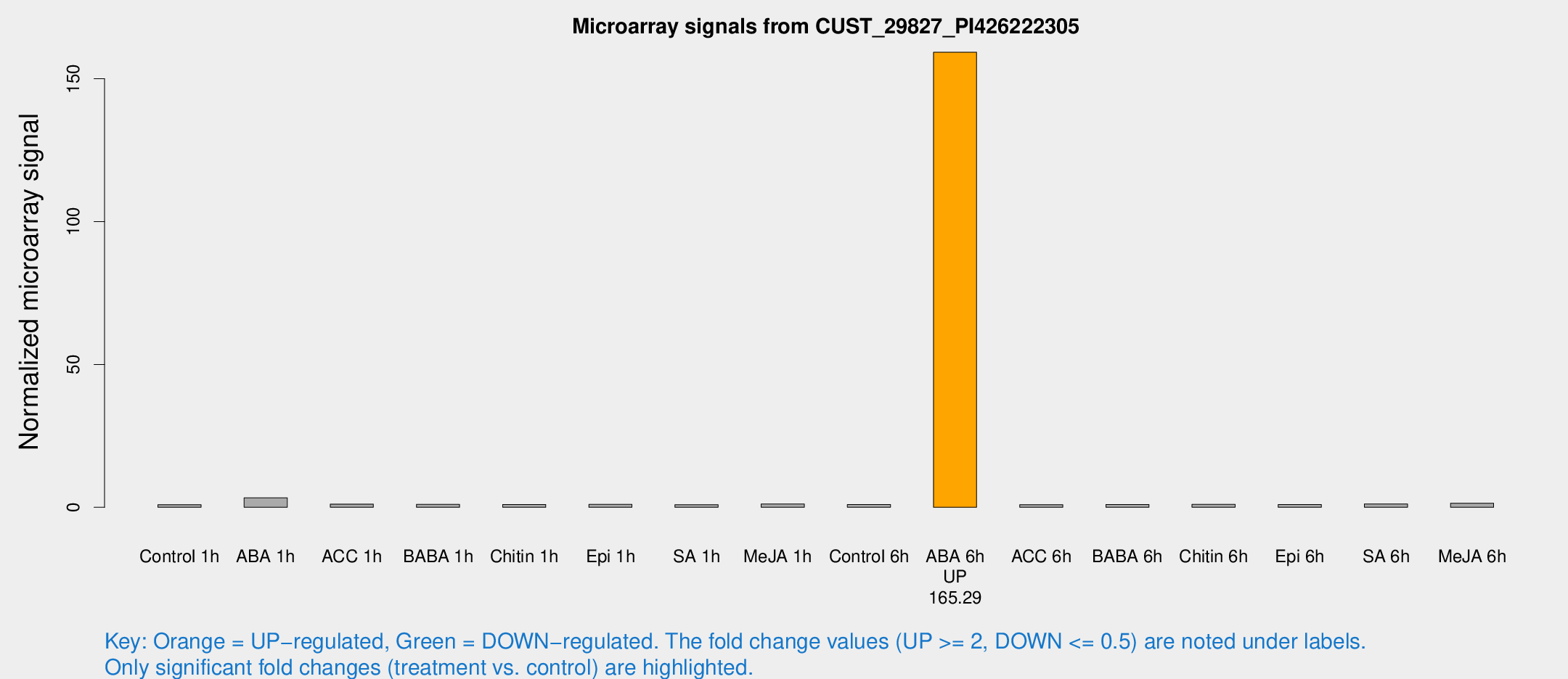
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.42052 | 3.7327 | 0.916202 | 0.530625 |
| ABA 1h | 23.0966 | 6.91165 | 3.31252 | 1.17302 |
| ACC 1h | 8.24844 | 4.78677 | 1.12629 | 0.633207 |
| BABA 1h | 6.79098 | 3.96085 | 1.00431 | 0.582945 |
| Chitin 1h | 6.31887 | 3.68784 | 0.998823 | 0.578406 |
| Epi 1h | 6.15722 | 3.58713 | 1.01284 | 0.586576 |
| SA 1h | 6.37829 | 3.70758 | 0.887272 | 0.514482 |
| Me-JA 1h | 6.57733 | 3.84222 | 1.14449 | 0.662675 |
| Control 6h | 6.75728 | 3.93 | 0.963606 | 0.559165 |
| ABA 6h | 1256.01 | 277.015 | 159.279 | 35.0283 |
| ACC 6h | 7.5692 | 4.48573 | 0.924446 | 0.535319 |
| BABA 6h | 7.9267 | 4.3695 | 1.00202 | 0.555979 |
| Chitin 6h | 7.50307 | 4.35428 | 1.00752 | 0.583378 |
| Epi 6h | 7.8538 | 4.65375 | 0.976214 | 0.565282 |
| SA 6h | 8.3638 | 4.16718 | 1.18999 | 0.614977 |
| Me-JA 6h | 11.1912 | 3.89331 | 1.44527 | 0.640046 |
Source Transcript PGSC0003DMT400033409 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G38540.1 | +2 | 6e-16 | 72 | 47/97 (48%) | lipid transfer protein 1 | chr2:16130418-16130893 FORWARD LENGTH=118 |
