Probe CUST_29788_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29788_PI426222305 | JHI_St_60k_v1 | DMT400053795 | GACAAACTTCAAAGTGTGTATCCTTATCTCATCAATGAAAGGCCAGCTTTTGTTTTAAAG |
All Microarray Probes Designed to Gene DMG400020870
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29778_PI426222305 | JHI_St_60k_v1 | DMT400053796 | GACAAACTTCAAAGTGTGTATCCTTATCTCATCAATGAAAGGCCAGCTTTTGTTTTAAAG |
| CUST_29788_PI426222305 | JHI_St_60k_v1 | DMT400053795 | GACAAACTTCAAAGTGTGTATCCTTATCTCATCAATGAAAGGCCAGCTTTTGTTTTAAAG |
Microarray Signals from CUST_29788_PI426222305
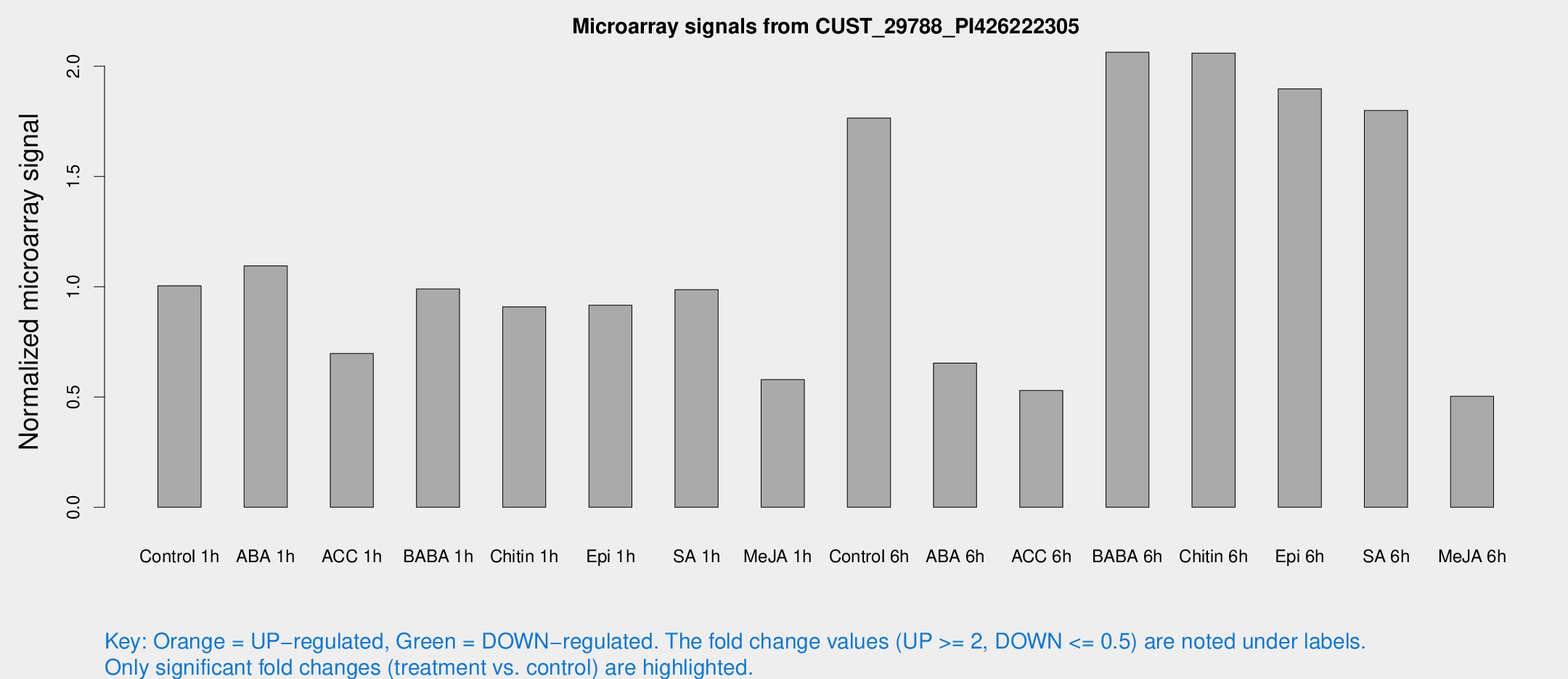
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 47.7864 | 15.1126 | 1.00444 | 0.338717 |
| ABA 1h | 41.5613 | 3.81245 | 1.09441 | 0.100712 |
| ACC 1h | 35.3537 | 12.9527 | 0.697686 | 0.249126 |
| BABA 1h | 42.3457 | 8.47407 | 0.99019 | 0.128223 |
| Chitin 1h | 45.9357 | 23.091 | 0.909098 | 0.547697 |
| Epi 1h | 34.6815 | 5.2394 | 0.916213 | 0.132936 |
| SA 1h | 43.9393 | 5.37696 | 0.98691 | 0.158691 |
| Me-JA 1h | 22.6615 | 6.66146 | 0.579398 | 0.231457 |
| Control 6h | 89.0639 | 30.2313 | 1.76499 | 0.687659 |
| ABA 6h | 30.4504 | 4.64493 | 0.653969 | 0.117551 |
| ACC 6h | 31.9887 | 11.5904 | 0.529829 | 0.258862 |
| BABA 6h | 100.333 | 12.9961 | 2.06325 | 0.217691 |
| Chitin 6h | 94.4207 | 9.68471 | 2.05912 | 0.162525 |
| Epi 6h | 98.1627 | 25.2264 | 1.89726 | 0.666108 |
| SA 6h | 78.1089 | 12.5174 | 1.79918 | 0.511279 |
| Me-JA 6h | 28.5103 | 11.2065 | 0.503128 | 0.342067 |
Source Transcript PGSC0003DMT400053795 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G53430.1 | +1 | 1e-22 | 55 | 20/38 (53%) | SET domain group 29 | chr5:21677623-21683166 FORWARD LENGTH=1043 |
