Probe CUST_29477_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29477_PI426222305 | JHI_St_60k_v1 | DMT400075870 | ATTCCGGAAATATTATCATGTAAAATTCTGCTATCACACTTTGGGAGGACTTCAAGATCA |
All Microarray Probes Designed to Gene DMG400029498
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_29477_PI426222305 | JHI_St_60k_v1 | DMT400075870 | ATTCCGGAAATATTATCATGTAAAATTCTGCTATCACACTTTGGGAGGACTTCAAGATCA |
| CUST_29418_PI426222305 | JHI_St_60k_v1 | DMT400075869 | GGACCACTTAAGATTTCTAGCTCCCAATAAAGTTTGTAACATTCAAATGCTCATCTATCA |
Microarray Signals from CUST_29477_PI426222305
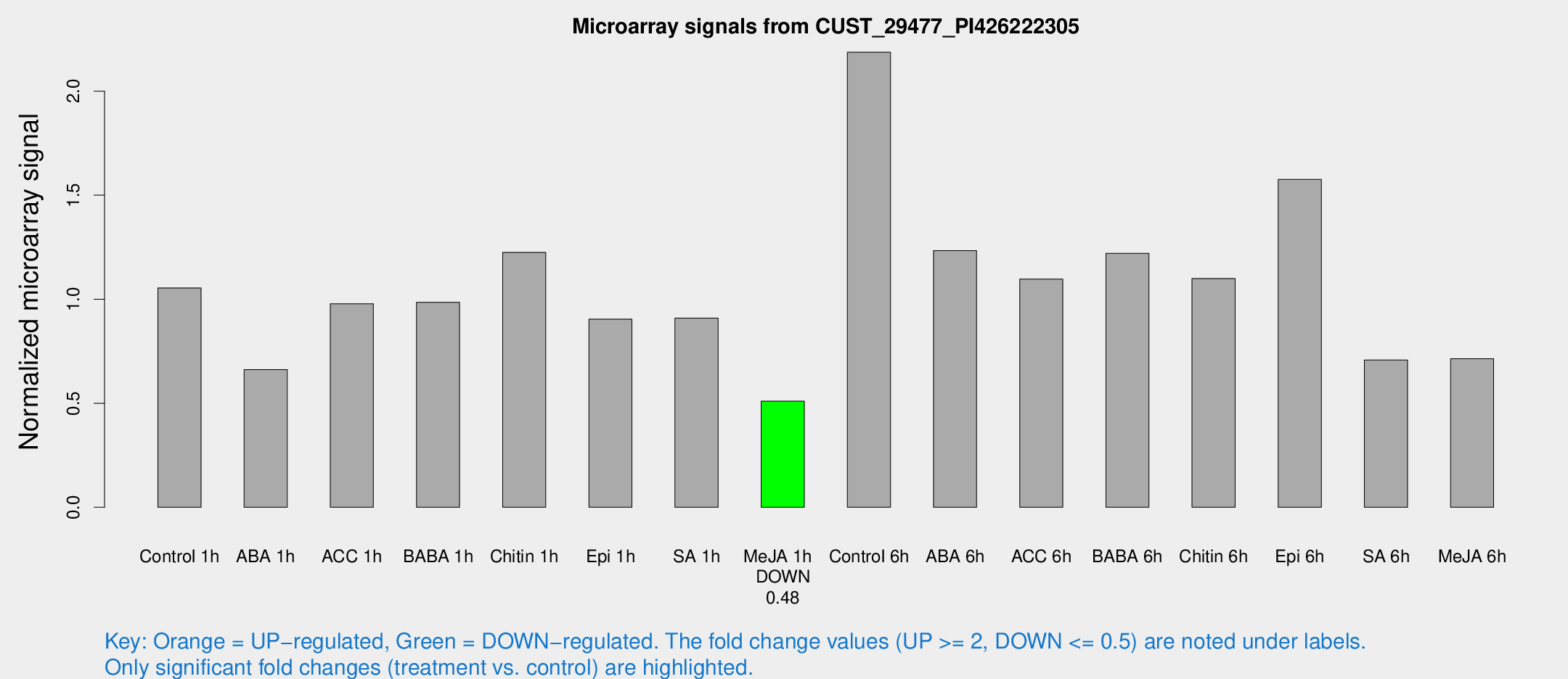
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 16.4066 | 3.63 | 1.0542 | 0.243222 |
| ABA 1h | 9.18569 | 3.26891 | 0.662221 | 0.26519 |
| ACC 1h | 16.0982 | 4.25227 | 0.978057 | 0.336831 |
| BABA 1h | 14.291 | 3.7578 | 0.985552 | 0.271252 |
| Chitin 1h | 16.4098 | 3.5076 | 1.22527 | 0.26196 |
| Epi 1h | 13.1863 | 4.24313 | 0.904349 | 0.36196 |
| SA 1h | 15.374 | 4.18462 | 0.909415 | 0.40832 |
| Me-JA 1h | 6.26113 | 3.48257 | 0.510025 | 0.282118 |
| Control 6h | 33.8181 | 6.62292 | 2.18708 | 0.32698 |
| ABA 6h | 21.9937 | 7.29908 | 1.2339 | 0.393813 |
| ACC 6h | 18.9535 | 4.34279 | 1.09691 | 0.259544 |
| BABA 6h | 25.1623 | 9.82373 | 1.22053 | 0.681877 |
| Chitin 6h | 17.967 | 4.04198 | 1.09961 | 0.265834 |
| Epi 6h | 27.8337 | 6.63258 | 1.57613 | 0.378997 |
| SA 6h | 11.2026 | 3.78281 | 0.707791 | 0.280089 |
| Me-JA 6h | 11.362 | 3.56135 | 0.714193 | 0.268026 |
Source Transcript PGSC0003DMT400075870 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G01910.1 | +1 | 0.0 | 680 | 383/622 (62%) | Microtubule associated protein (MAP65/ASE1) family protein | chr2:417191-420182 FORWARD LENGTH=608 |
