Probe CUST_28112_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28112_PI426222305 | JHI_St_60k_v1 | DMT400004191 | CCAGCAGAACCTCAAGAAGAATGATATGTACCTAGCTATTCTTTGCTTTAGTAAAATTAC |
All Microarray Probes Designed to Gene DMG400001671
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_28055_PI426222305 | JHI_St_60k_v1 | DMT400004192 | AGAGCTTGCTGATAATAGCAGTCGAAACATACTGCATACGAAAGAATTTAGGTAAGTTAT |
| CUST_28112_PI426222305 | JHI_St_60k_v1 | DMT400004191 | CCAGCAGAACCTCAAGAAGAATGATATGTACCTAGCTATTCTTTGCTTTAGTAAAATTAC |
Microarray Signals from CUST_28112_PI426222305
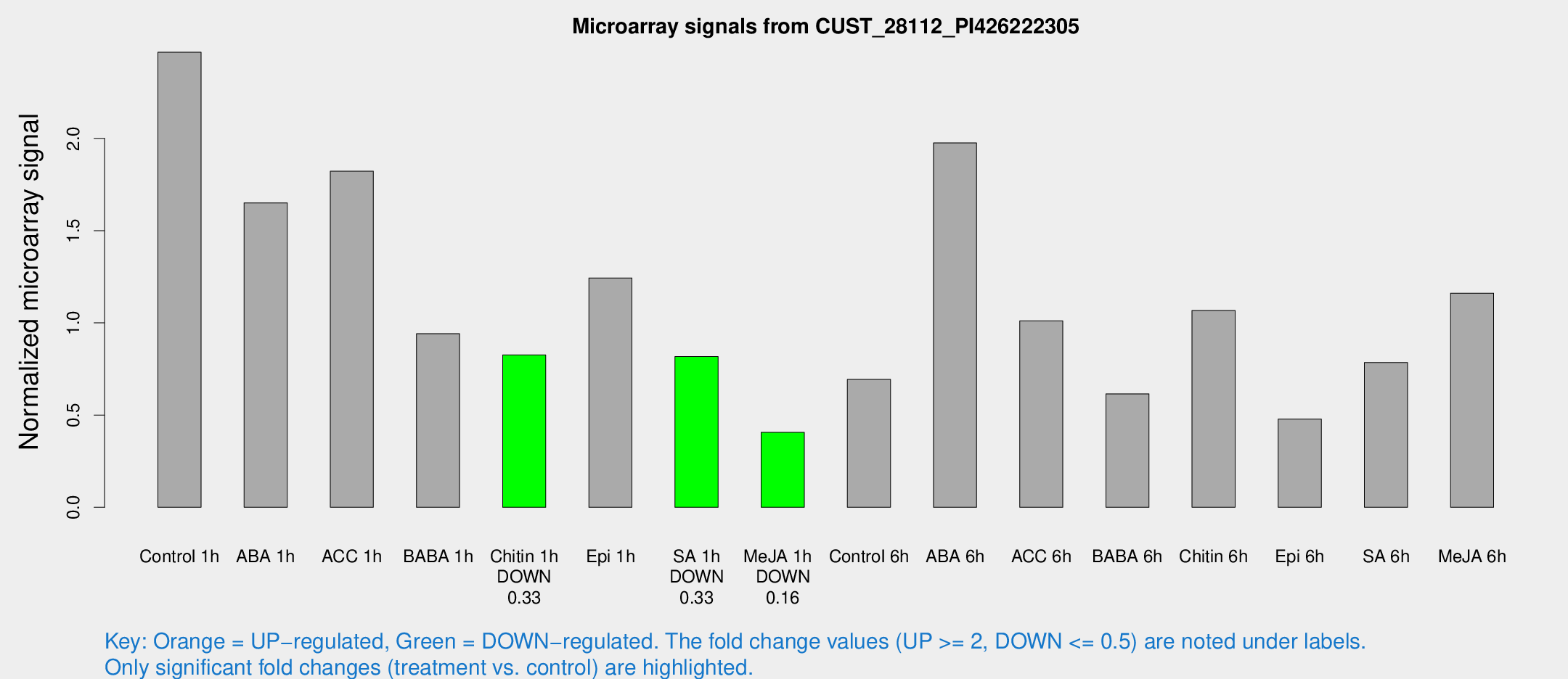
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 2080.97 | 304.858 | 2.46754 | 0.283519 |
| ABA 1h | 1248.7 | 252.827 | 1.65087 | 0.375832 |
| ACC 1h | 1568.68 | 200.824 | 1.82245 | 0.115374 |
| BABA 1h | 854.097 | 260.156 | 0.941531 | 0.292708 |
| Chitin 1h | 627.272 | 94.1295 | 0.825986 | 0.0861204 |
| Epi 1h | 890.217 | 62.8487 | 1.24361 | 0.097935 |
| SA 1h | 725.158 | 156.193 | 0.81749 | 0.131827 |
| Me-JA 1h | 286.7 | 57.8658 | 0.406077 | 0.0698882 |
| Control 6h | 592.126 | 114.562 | 0.693212 | 0.135785 |
| ABA 6h | 1823.34 | 385.233 | 1.97556 | 0.415662 |
| ACC 6h | 969.931 | 127.139 | 1.01079 | 0.234712 |
| BABA 6h | 589.971 | 117.707 | 0.615379 | 0.135759 |
| Chitin 6h | 945.34 | 97.3593 | 1.06724 | 0.130122 |
| Epi 6h | 452.266 | 64.8805 | 0.477911 | 0.0352003 |
| SA 6h | 666.956 | 129.461 | 0.784729 | 0.0792784 |
| Me-JA 6h | 1055.99 | 307.083 | 1.16114 | 0.424666 |
Source Transcript PGSC0003DMT400004191 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G32450.1 | +3 | 0.0 | 842 | 429/602 (71%) | nitrate transporter 1.5 | chr1:11715337-11719807 REVERSE LENGTH=614 |
