Probe CUST_27105_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27105_PI426222305 | JHI_St_60k_v1 | DMT400026310 | TTATTAAGAAGATCAACCATGAGAGATTGCTGCAGAGGCAAGAATCCGATCAGCGTGCCT |
All Microarray Probes Designed to Gene DMG400010154
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_27128_PI426222305 | JHI_St_60k_v1 | DMT400026311 | GTACAACAGTTACCTTGGTTGTATCTGTAAAGTTTATCACTTTCAGTATTTCCAGTGAGA |
| CUST_27105_PI426222305 | JHI_St_60k_v1 | DMT400026310 | TTATTAAGAAGATCAACCATGAGAGATTGCTGCAGAGGCAAGAATCCGATCAGCGTGCCT |
Microarray Signals from CUST_27105_PI426222305
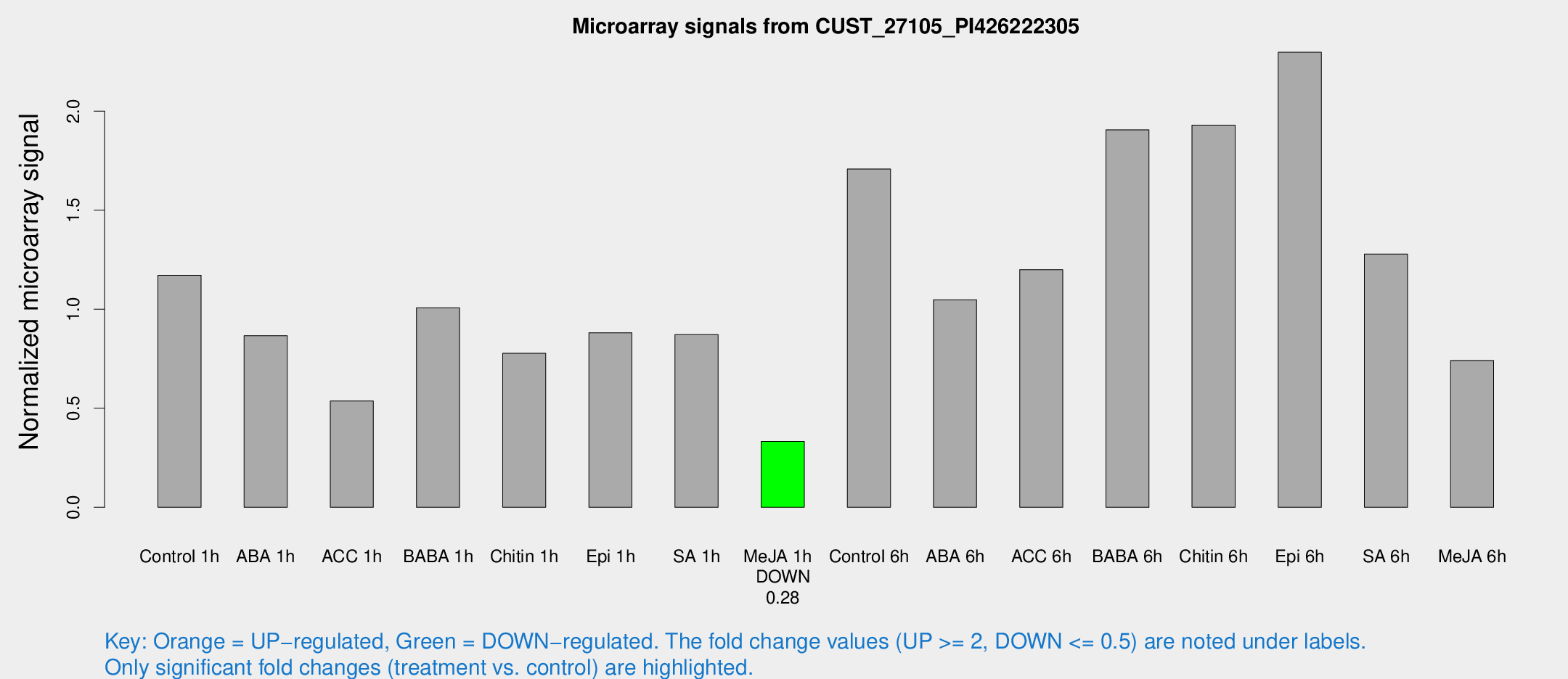
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 116.734 | 29.4821 | 1.17139 | 0.212184 |
| ABA 1h | 87.0665 | 39.2069 | 0.866152 | 0.343651 |
| ACC 1h | 64.5124 | 23.4369 | 0.536711 | 0.28198 |
| BABA 1h | 101.977 | 30.3398 | 1.00719 | 0.268403 |
| Chitin 1h | 68.7206 | 13.9526 | 0.777409 | 0.116189 |
| Epi 1h | 73.3576 | 11.2288 | 0.881252 | 0.123263 |
| SA 1h | 87.1654 | 17.2684 | 0.871712 | 0.135479 |
| Me-JA 1h | 25.5001 | 3.307 | 0.332401 | 0.0429793 |
| Control 6h | 170.754 | 38.0168 | 1.70821 | 0.294446 |
| ABA 6h | 106.944 | 16.5016 | 1.0482 | 0.204251 |
| ACC 6h | 135.438 | 28.1546 | 1.19985 | 0.155294 |
| BABA 6h | 200.261 | 12.0384 | 1.90624 | 0.114554 |
| Chitin 6h | 193.661 | 13.791 | 1.92958 | 0.127669 |
| Epi 6h | 246.701 | 30.101 | 2.29792 | 0.234475 |
| SA 6h | 128.337 | 32.7444 | 1.2785 | 0.233303 |
| Me-JA 6h | 71.8384 | 13.0802 | 0.741384 | 0.0828303 |
Source Transcript PGSC0003DMT400026310 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
