Probe CUST_26384_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26384_PI426222305 | JHI_St_60k_v1 | DMT400037110 | TAGCTTCATGGATATGTGTAATTAGCCTCATGGCACTTGTATTAATGAGTTCTGTTAGGG |
All Microarray Probes Designed to Gene DMG400014304
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_26384_PI426222305 | JHI_St_60k_v1 | DMT400037110 | TAGCTTCATGGATATGTGTAATTAGCCTCATGGCACTTGTATTAATGAGTTCTGTTAGGG |
| CUST_26368_PI426222305 | JHI_St_60k_v1 | DMT400037109 | GATGATGTTTTTCCTGGACTTGTCATTAAGATTGTTCCTTTCAACAATAACAAGTCATGG |
Microarray Signals from CUST_26384_PI426222305
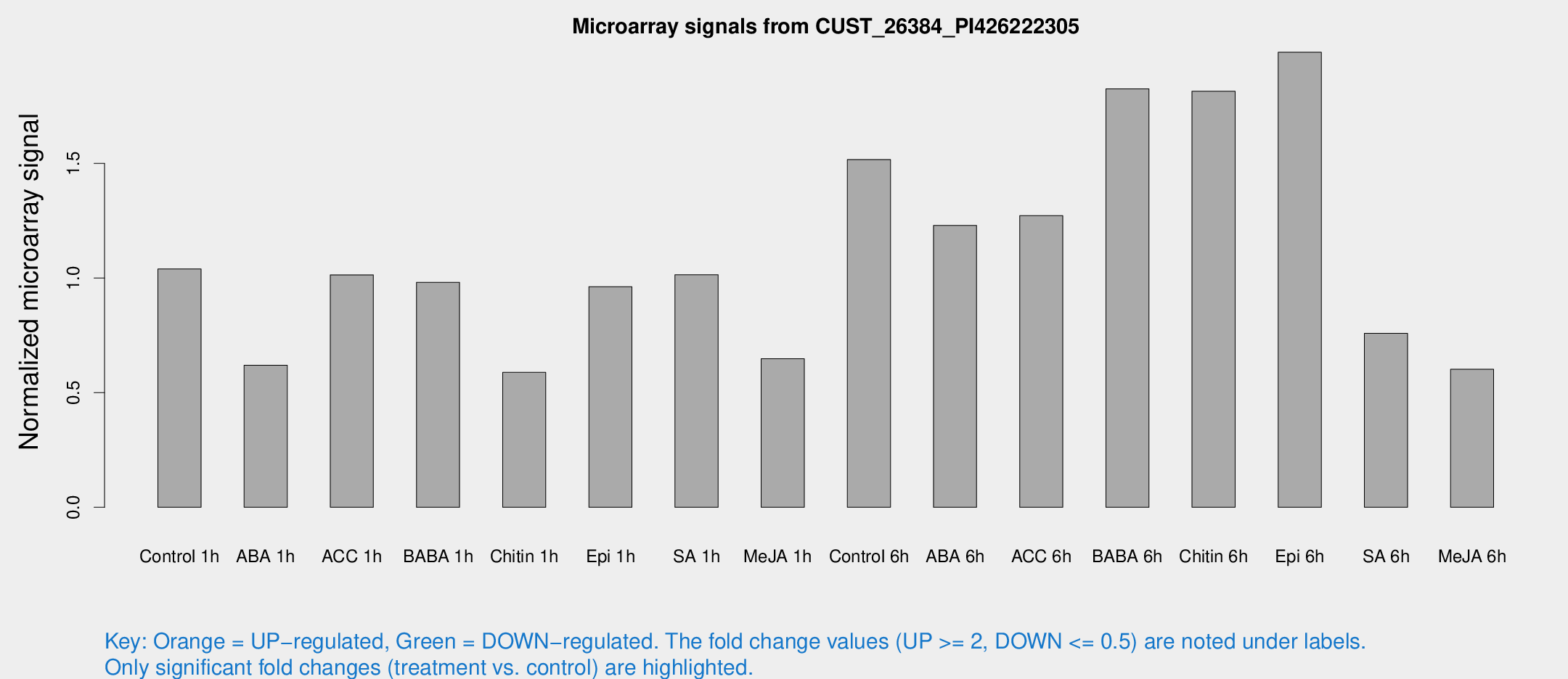
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 55.4968 | 10.0542 | 1.03972 | 0.120878 |
| ABA 1h | 28.6739 | 3.78807 | 0.61934 | 0.0828463 |
| ACC 1h | 60.9573 | 17.9671 | 1.01389 | 0.324125 |
| BABA 1h | 50.5051 | 8.46973 | 0.981338 | 0.0977662 |
| Chitin 1h | 27.6616 | 3.84998 | 0.588418 | 0.0822185 |
| Epi 1h | 43.5109 | 4.31517 | 0.962285 | 0.0977803 |
| SA 1h | 56.1262 | 10.562 | 1.01405 | 0.245038 |
| Me-JA 1h | 27.7639 | 3.94573 | 0.647746 | 0.093676 |
| Control 6h | 81.1805 | 13.2623 | 1.51681 | 0.127944 |
| ABA 6h | 67.792 | 5.52262 | 1.22938 | 0.100095 |
| ACC 6h | 80.6404 | 21.2752 | 1.27233 | 0.16971 |
| BABA 6h | 113.034 | 25.7593 | 1.82545 | 0.443674 |
| Chitin 6h | 102.004 | 14.5932 | 1.81462 | 0.247493 |
| Epi 6h | 116.918 | 10.4961 | 1.98493 | 0.332013 |
| SA 6h | 40.397 | 7.60048 | 0.758524 | 0.094851 |
| Me-JA 6h | 35.8686 | 11.334 | 0.602018 | 0.225215 |
Source Transcript PGSC0003DMT400037110 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G62150.1 | +3 | 2e-39 | 133 | 62/98 (63%) | peptidoglycan-binding LysM domain-containing protein | chr5:24958325-24958633 FORWARD LENGTH=102 |
