Probe CUST_25643_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25643_PI426222305 | JHI_St_60k_v1 | DMT400037336 | ATTTGCTCGATTTCTTCTATCGTCGTTGAAGTTTAGCAGCAGCATCGCAGATCTGTAATG |
All Microarray Probes Designed to Gene DMG400014405
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25579_PI426222305 | JHI_St_60k_v1 | DMT400037335 | TGCGAAATGCCTGATTATGCTTTGGTTATATATTGAAGGCTTTGGACAAGATTCGCTTTG |
| CUST_25643_PI426222305 | JHI_St_60k_v1 | DMT400037336 | ATTTGCTCGATTTCTTCTATCGTCGTTGAAGTTTAGCAGCAGCATCGCAGATCTGTAATG |
Microarray Signals from CUST_25643_PI426222305
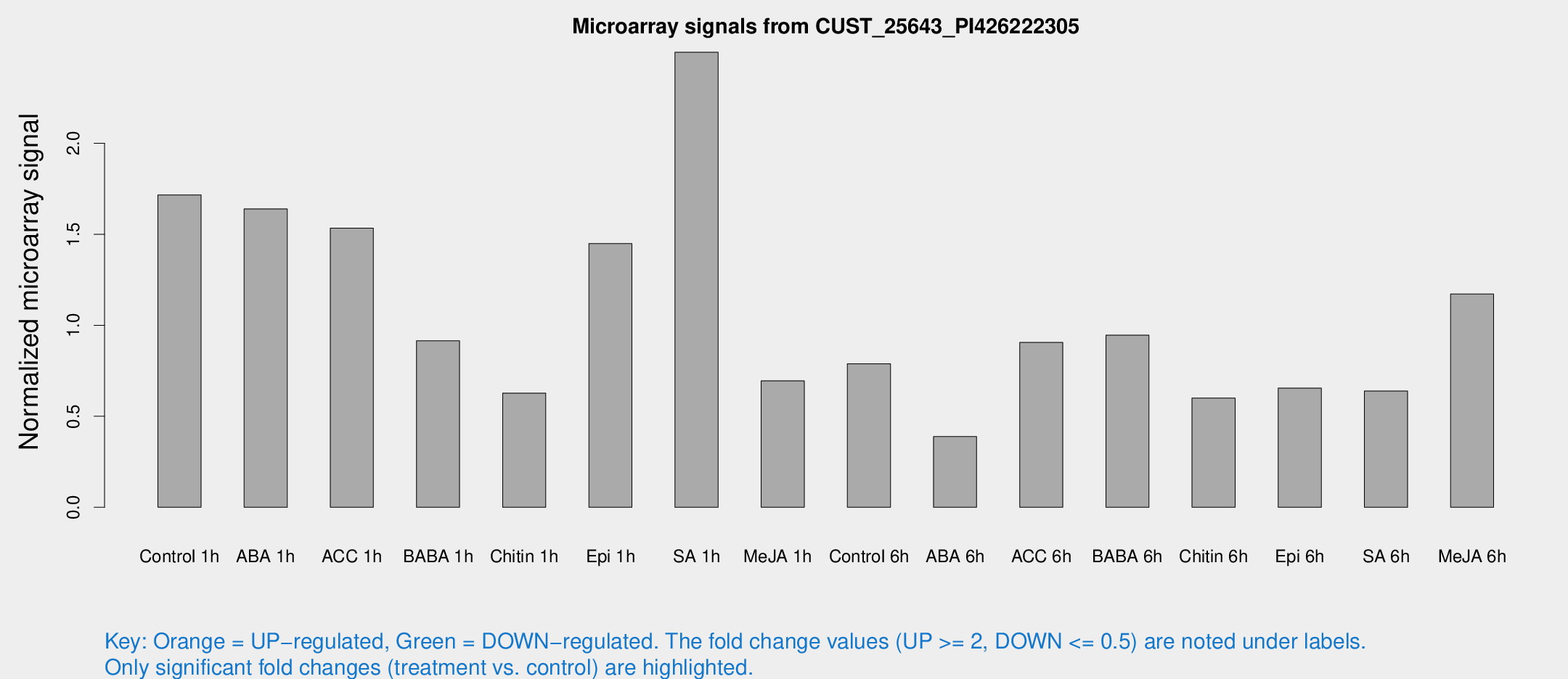
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 61.8914 | 12 | 1.71622 | 0.438056 |
| ABA 1h | 54.5136 | 15.7457 | 1.63947 | 0.506797 |
| ACC 1h | 57.0147 | 10.3263 | 1.53375 | 0.197338 |
| BABA 1h | 33.538 | 10.3312 | 0.914544 | 0.269244 |
| Chitin 1h | 20.8995 | 4.75074 | 0.627127 | 0.219706 |
| Epi 1h | 47.244 | 12.9779 | 1.44876 | 0.471192 |
| SA 1h | 92.9513 | 19.029 | 2.4998 | 0.649869 |
| Me-JA 1h | 21.6275 | 6.35774 | 0.694929 | 0.305595 |
| Control 6h | 30.7876 | 10.8323 | 0.788745 | 0.22954 |
| ABA 6h | 19.1481 | 10.2727 | 0.388825 | 0.26903 |
| ACC 6h | 36.4355 | 5.15067 | 0.905888 | 0.170906 |
| BABA 6h | 37.4953 | 4.73363 | 0.945607 | 0.124696 |
| Chitin 6h | 24.7057 | 6.86092 | 0.600108 | 0.230934 |
| Epi 6h | 25.9337 | 4.74666 | 0.654653 | 0.124031 |
| SA 6h | 28.441 | 13.2106 | 0.638645 | 0.315381 |
| Me-JA 6h | 42.1937 | 8.02792 | 1.17144 | 0.349031 |
Source Transcript PGSC0003DMT400037336 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G56010.1 | +2 | 0.0 | 1112 | 652/700 (93%) | heat shock protein 81-3 | chr5:22681410-22683911 FORWARD LENGTH=699 |
