Probe CUST_25415_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25415_PI426222305 | JHI_St_60k_v1 | DMT400043708 | AAAGGATTGTTATTCCCAAAGGGAGGTTGGGAAGAAGATGAAACGATTGAGGATGCAGCG |
All Microarray Probes Designed to Gene DMG400016977
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25415_PI426222305 | JHI_St_60k_v1 | DMT400043708 | AAAGGATTGTTATTCCCAAAGGGAGGTTGGGAAGAAGATGAAACGATTGAGGATGCAGCG |
| CUST_25463_PI426222305 | JHI_St_60k_v1 | DMT400043710 | CAATCTGTAGATGAATGTTCGCGAAGCAAGAAAACAATGCCAAAATGGATGGATGAAAGA |
Microarray Signals from CUST_25415_PI426222305
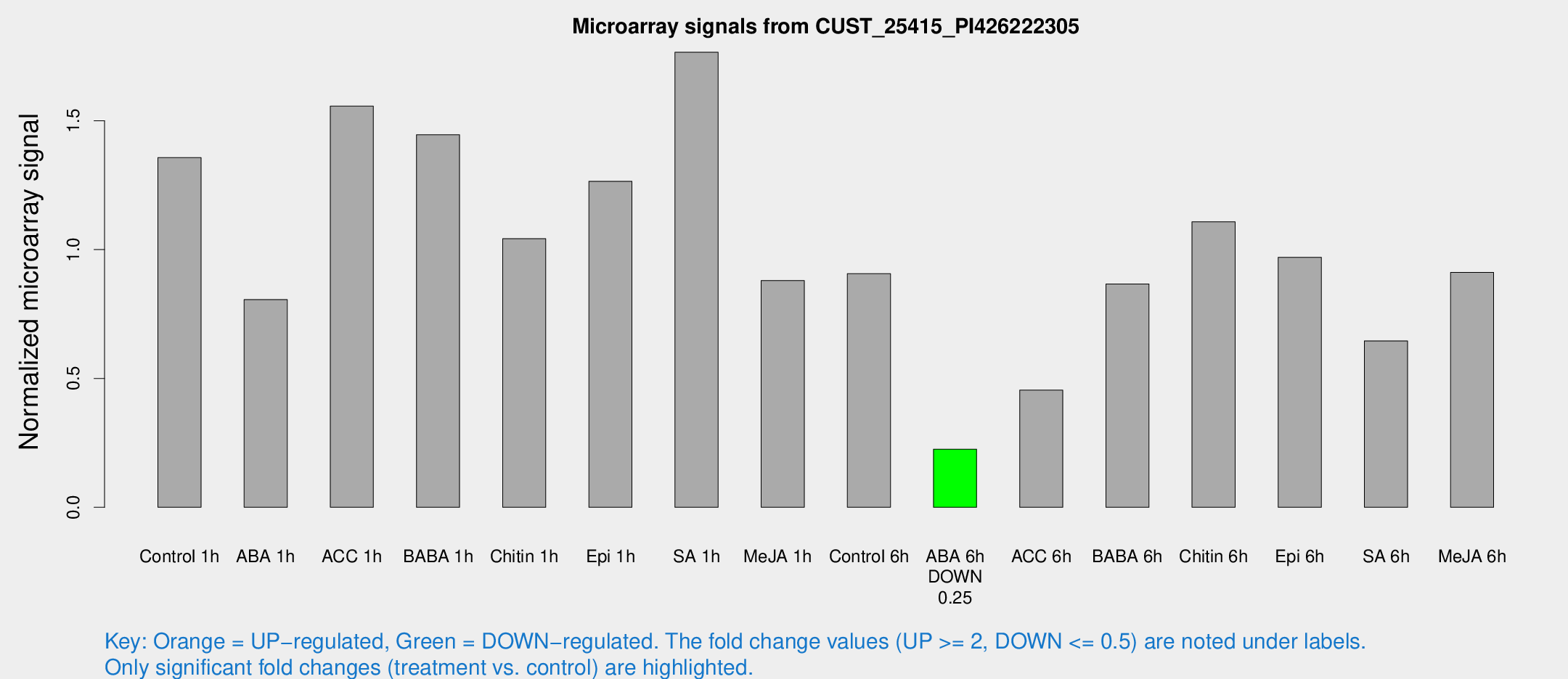
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 224.798 | 46.9704 | 1.35698 | 0.194507 |
| ABA 1h | 115.372 | 14.5252 | 0.806268 | 0.0879207 |
| ACC 1h | 280.802 | 77.7168 | 1.55653 | 0.413857 |
| BABA 1h | 222.345 | 13.3041 | 1.44568 | 0.0863532 |
| Chitin 1h | 154.151 | 28.0501 | 1.04245 | 0.10565 |
| Epi 1h | 175.246 | 12.5759 | 1.26505 | 0.0801676 |
| SA 1h | 293.846 | 39.8553 | 1.76587 | 0.13089 |
| Me-JA 1h | 116.634 | 15.7894 | 0.879959 | 0.12819 |
| Control 6h | 161.376 | 48.1375 | 0.906452 | 0.252644 |
| ABA 6h | 39.4924 | 6.67014 | 0.225699 | 0.0336133 |
| ACC 6h | 89.3997 | 22.7756 | 0.454488 | 0.0838152 |
| BABA 6h | 166.595 | 43.8342 | 0.866706 | 0.230413 |
| Chitin 6h | 188.305 | 11.4904 | 1.10792 | 0.0682827 |
| Epi 6h | 182.474 | 37.6405 | 0.970253 | 0.277413 |
| SA 6h | 112.811 | 30.9665 | 0.645854 | 0.151598 |
| Me-JA 6h | 161.427 | 54.5673 | 0.911242 | 0.23747 |
Source Transcript PGSC0003DMT400043708 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G01670.1 | +3 | 7e-62 | 199 | 99/170 (58%) | nudix hydrolase homolog 17 | chr2:296889-297818 REVERSE LENGTH=182 |
