Probe CUST_24840_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24840_PI426222305 | JHI_St_60k_v1 | DMT400045130 | CGCGAAGTCAATTCTAAGCATAACATTGTTACTGAAGAATGAAGTATGTAGTGATTGATC |
All Microarray Probes Designed to Gene DMG400017505
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24848_PI426222305 | JHI_St_60k_v1 | DMT400045129 | CGCGAAGTCAATTCTAAGCATAACATTGTTACTGAAGAATGAAGTATGTAGTGATTGATC |
| CUST_24840_PI426222305 | JHI_St_60k_v1 | DMT400045130 | CGCGAAGTCAATTCTAAGCATAACATTGTTACTGAAGAATGAAGTATGTAGTGATTGATC |
Microarray Signals from CUST_24840_PI426222305
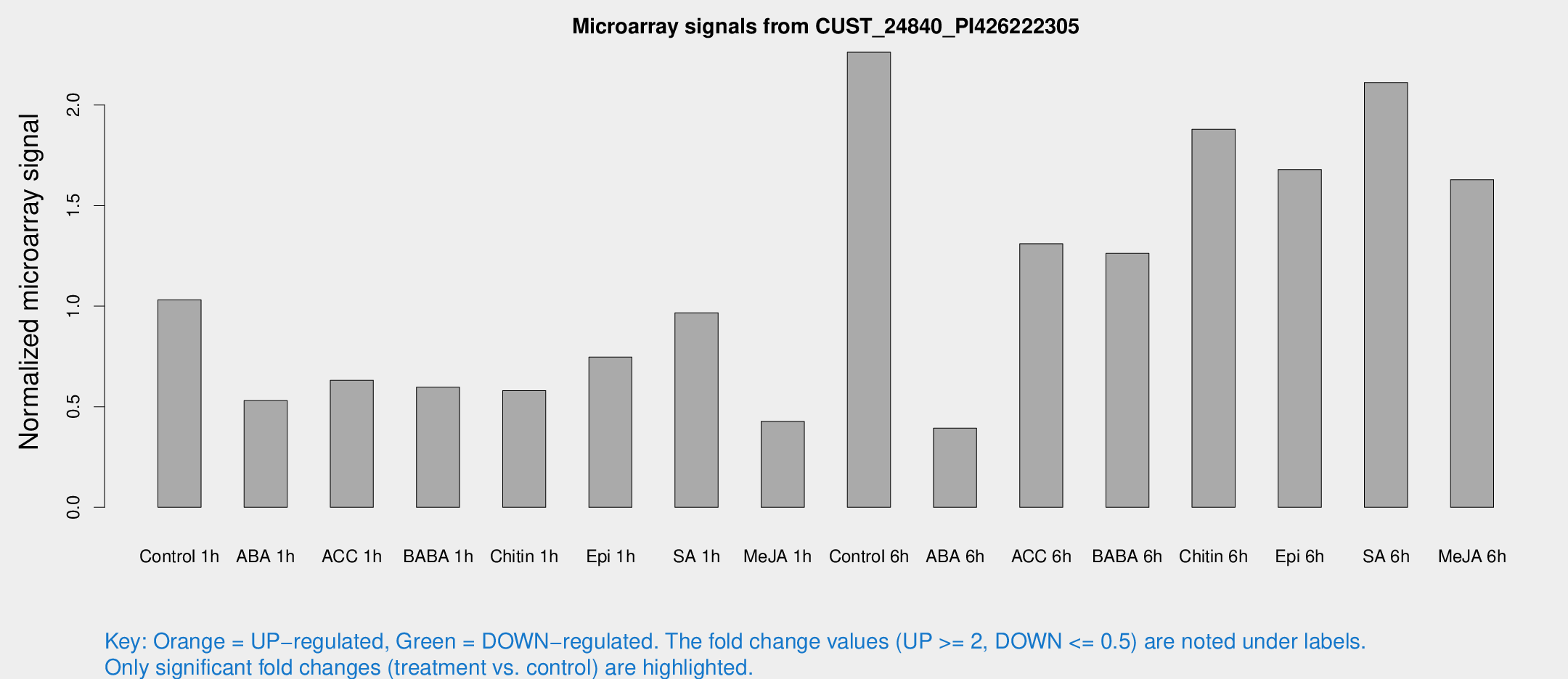
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 105.694 | 19.8667 | 1.03142 | 0.151063 |
| ABA 1h | 46.8175 | 4.41854 | 0.530439 | 0.0829699 |
| ACC 1h | 71.4324 | 21.9281 | 0.631298 | 0.180392 |
| BABA 1h | 63.5278 | 22.02 | 0.596925 | 0.179543 |
| Chitin 1h | 54.895 | 14.1295 | 0.58027 | 0.101455 |
| Epi 1h | 64.9713 | 8.1395 | 0.746455 | 0.0957978 |
| SA 1h | 106.098 | 25.5065 | 0.967253 | 0.238692 |
| Me-JA 1h | 34.6573 | 4.09819 | 0.426604 | 0.0499785 |
| Control 6h | 241.6 | 63.4491 | 2.26241 | 0.476526 |
| ABA 6h | 42.6435 | 7.34977 | 0.392983 | 0.0631953 |
| ACC 6h | 153.119 | 25.3986 | 1.31037 | 0.27307 |
| BABA 6h | 154.14 | 45.6903 | 1.26225 | 0.461171 |
| Chitin 6h | 207.936 | 46.9799 | 1.87964 | 0.370617 |
| Epi 6h | 190.162 | 21.249 | 1.67933 | 0.10462 |
| SA 6h | 219.059 | 47.1064 | 2.11144 | 0.279053 |
| Me-JA 6h | 210.364 | 111.608 | 1.62889 | 0.858121 |
Source Transcript PGSC0003DMT400045130 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G17980.1 | +1 | 3e-89 | 279 | 142/249 (57%) | NAC domain containing protein 71 | chr4:9978850-9980038 REVERSE LENGTH=262 |
