Probe CUST_24822_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24822_PI426222305 | JHI_St_60k_v1 | DMT400045227 | ATTTGATGGTTTGCTTTTGCCATGAACGTTCATTTCGACCTCCTAGGGTGTCAACAAAGG |
All Microarray Probes Designed to Gene DMG401017546
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24822_PI426222305 | JHI_St_60k_v1 | DMT400045227 | ATTTGATGGTTTGCTTTTGCCATGAACGTTCATTTCGACCTCCTAGGGTGTCAACAAAGG |
| CUST_24868_PI426222305 | JHI_St_60k_v1 | DMT400045229 | TGCAGCAGAGCATGGCTATTTTGTGAGGCCAAATCATGATGCCAAATGGGAAACTTGTGT |
Microarray Signals from CUST_24822_PI426222305
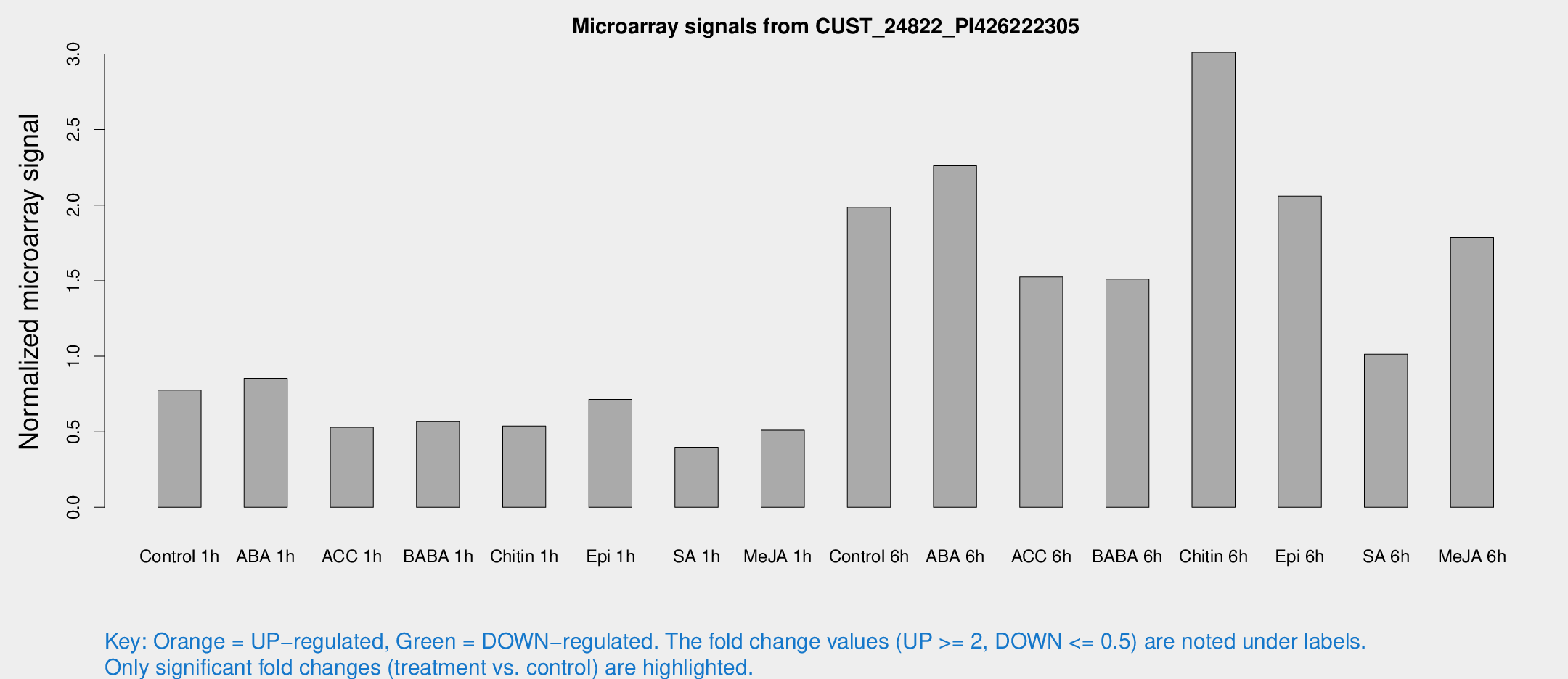
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 12.0251 | 3.6996 | 0.776182 | 0.240458 |
| ABA 1h | 12.8706 | 3.77013 | 0.853249 | 0.389572 |
| ACC 1h | 8.57515 | 4.58323 | 0.530219 | 0.278714 |
| BABA 1h | 8.89081 | 3.86626 | 0.567456 | 0.276016 |
| Chitin 1h | 7.59204 | 3.60842 | 0.538202 | 0.268293 |
| Epi 1h | 9.92735 | 3.54748 | 0.714413 | 0.283611 |
| SA 1h | 6.28967 | 3.64663 | 0.397085 | 0.229949 |
| Me-JA 1h | 6.45909 | 3.76153 | 0.511006 | 0.295977 |
| Control 6h | 42.0684 | 24.1737 | 1.98512 | 1.2356 |
| ABA 6h | 42.1113 | 12.5839 | 2.26026 | 0.854657 |
| ACC 6h | 27.0337 | 4.67701 | 1.5242 | 0.310538 |
| BABA 6h | 27.9629 | 7.69805 | 1.51053 | 0.388148 |
| Chitin 6h | 50.1057 | 5.79303 | 3.01183 | 0.37452 |
| Epi 6h | 37.4648 | 8.4596 | 2.05959 | 0.46238 |
| SA 6h | 19.5657 | 9.46376 | 1.01294 | 0.580802 |
| Me-JA 6h | 34.3726 | 16.8156 | 1.78473 | 0.778548 |
Source Transcript PGSC0003DMT400045227 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G17770.1 | +3 | 3e-32 | 124 | 69/89 (78%) | trehalose phosphatase/synthase 5 | chr4:9877055-9880084 FORWARD LENGTH=862 |
