Probe CUST_2476_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2476_PI426222305 | JHI_St_60k_v1 | DMT400027157 | AGGGAGAGTGATGTTGTAGTAAAGGGTTCAAAATTCAAGAGGATTTGTGTGTTTTGTGGA |
All Microarray Probes Designed to Gene DMG400010477
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2480_PI426222305 | JHI_St_60k_v1 | DMT400027158 | CAAGACACTCATGCCTCGAGAGGGTGTTTTGTCTTATGGAAGGTGTCAAGGAATATTGAT |
| CUST_2476_PI426222305 | JHI_St_60k_v1 | DMT400027157 | AGGGAGAGTGATGTTGTAGTAAAGGGTTCAAAATTCAAGAGGATTTGTGTGTTTTGTGGA |
Microarray Signals from CUST_2476_PI426222305
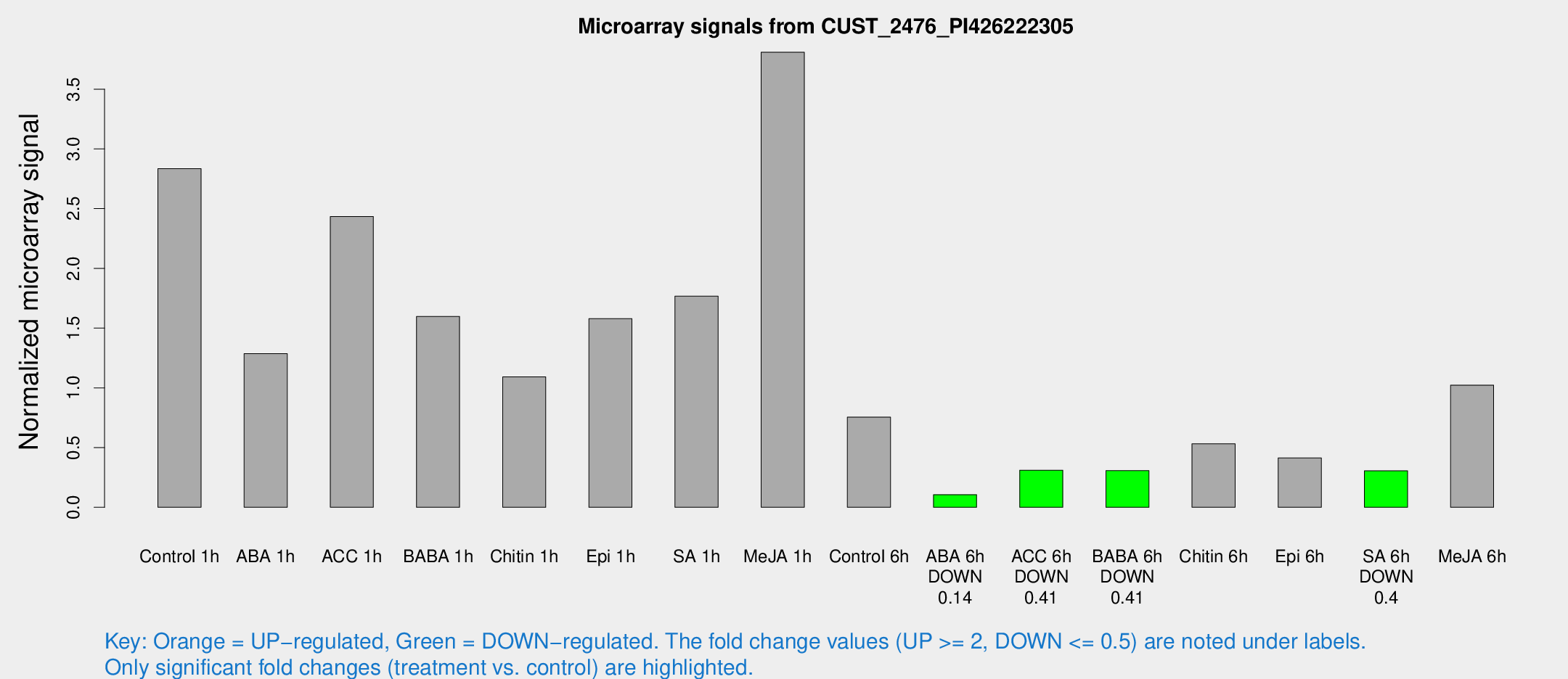
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 651.56 | 166.332 | 2.83444 | 0.513061 |
| ABA 1h | 252.426 | 36.4462 | 1.28667 | 0.142511 |
| ACC 1h | 580.671 | 156.103 | 2.43422 | 0.538259 |
| BABA 1h | 340.466 | 49.177 | 1.59763 | 0.137871 |
| Chitin 1h | 214.978 | 23.6306 | 1.09122 | 0.153244 |
| Epi 1h | 296.27 | 17.4702 | 1.5787 | 0.0929833 |
| SA 1h | 468.962 | 203.633 | 1.7675 | 0.859277 |
| Me-JA 1h | 738.425 | 206.303 | 3.80922 | 0.915355 |
| Control 6h | 170.229 | 31.796 | 0.754244 | 0.093488 |
| ABA 6h | 26.4359 | 7.48308 | 0.105924 | 0.0251182 |
| ACC 6h | 79.0301 | 12.3462 | 0.310924 | 0.0242512 |
| BABA 6h | 75.443 | 9.16349 | 0.306797 | 0.0327309 |
| Chitin 6h | 122.938 | 8.36127 | 0.53159 | 0.0361025 |
| Epi 6h | 108.152 | 25.9204 | 0.413202 | 0.148304 |
| SA 6h | 65.9556 | 6.05785 | 0.305058 | 0.0291966 |
| Me-JA 6h | 221.072 | 13.338 | 1.02264 | 0.0827758 |
Source Transcript PGSC0003DMT400027157 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G53450.1 | +2 | 6e-126 | 367 | 185/212 (87%) | Putative lysine decarboxylase family protein | chr3:19812977-19815430 REVERSE LENGTH=215 |
