Probe CUST_24664_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24664_PI426222305 | JHI_St_60k_v1 | DMT400054511 | GCACATAGGTGCACTCGATGATTGCCCATTTTTTAAAAAATTAATCAATTTGGTCCCATT |
All Microarray Probes Designed to Gene DMG400021157
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24664_PI426222305 | JHI_St_60k_v1 | DMT400054511 | GCACATAGGTGCACTCGATGATTGCCCATTTTTTAAAAAATTAATCAATTTGGTCCCATT |
Microarray Signals from CUST_24664_PI426222305
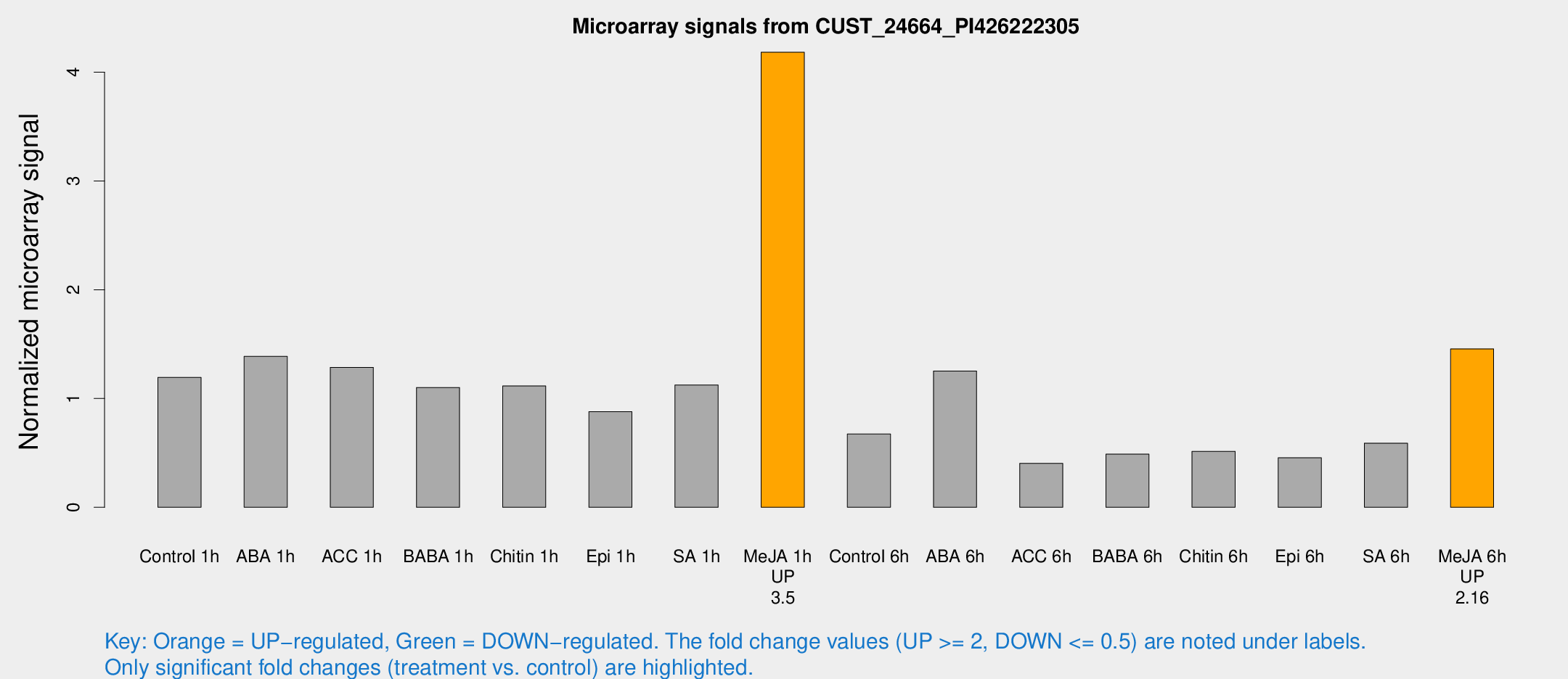
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1212.28 | 190.923 | 1.19441 | 0.123173 |
| ABA 1h | 1241.75 | 178.589 | 1.38812 | 0.102031 |
| ACC 1h | 1371.85 | 306.863 | 1.28641 | 0.239034 |
| BABA 1h | 1073.09 | 141.475 | 1.10128 | 0.0636902 |
| Chitin 1h | 1011.39 | 135.1 | 1.11522 | 0.160708 |
| Epi 1h | 757.691 | 48.2387 | 0.878922 | 0.0508903 |
| SA 1h | 1243.71 | 366.201 | 1.12487 | 0.365451 |
| Me-JA 1h | 3406.41 | 250.283 | 4.18303 | 0.241542 |
| Control 6h | 711.23 | 178.187 | 0.673404 | 0.115262 |
| ABA 6h | 1347.6 | 195.077 | 1.25305 | 0.10825 |
| ACC 6h | 468.726 | 67.1059 | 0.404127 | 0.100189 |
| BABA 6h | 544.314 | 33.7793 | 0.488383 | 0.0503107 |
| Chitin 6h | 561.677 | 105.643 | 0.514643 | 0.100509 |
| Epi 6h | 524.216 | 83.7439 | 0.455491 | 0.110541 |
| SA 6h | 592.433 | 94.0537 | 0.589809 | 0.0688538 |
| Me-JA 6h | 1439.95 | 83.3046 | 1.4561 | 0.174048 |
Source Transcript PGSC0003DMT400054511 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G34500.1 | +1 | 0.0 | 625 | 299/489 (61%) | cytochrome P450, family 710, subfamily A, polypeptide 1 | chr2:14539712-14541199 REVERSE LENGTH=495 |
