Probe CUST_23409_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23409_PI426222305 | JHI_St_60k_v1 | DMT400065697 | CTCATCTGCAGATAGAAACTGCGATAGAATTGAATTCGATGAATCTTTACAGGACTAATT |
All Microarray Probes Designed to Gene DMG400025559
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23409_PI426222305 | JHI_St_60k_v1 | DMT400065697 | CTCATCTGCAGATAGAAACTGCGATAGAATTGAATTCGATGAATCTTTACAGGACTAATT |
| CUST_23278_PI426222305 | JHI_St_60k_v1 | DMT400065696 | GCTGGTTGGGATGGAAAGATTCGGACATTCCATAACTATGGATTGCCTGTCCGAGTTTAA |
Microarray Signals from CUST_23409_PI426222305
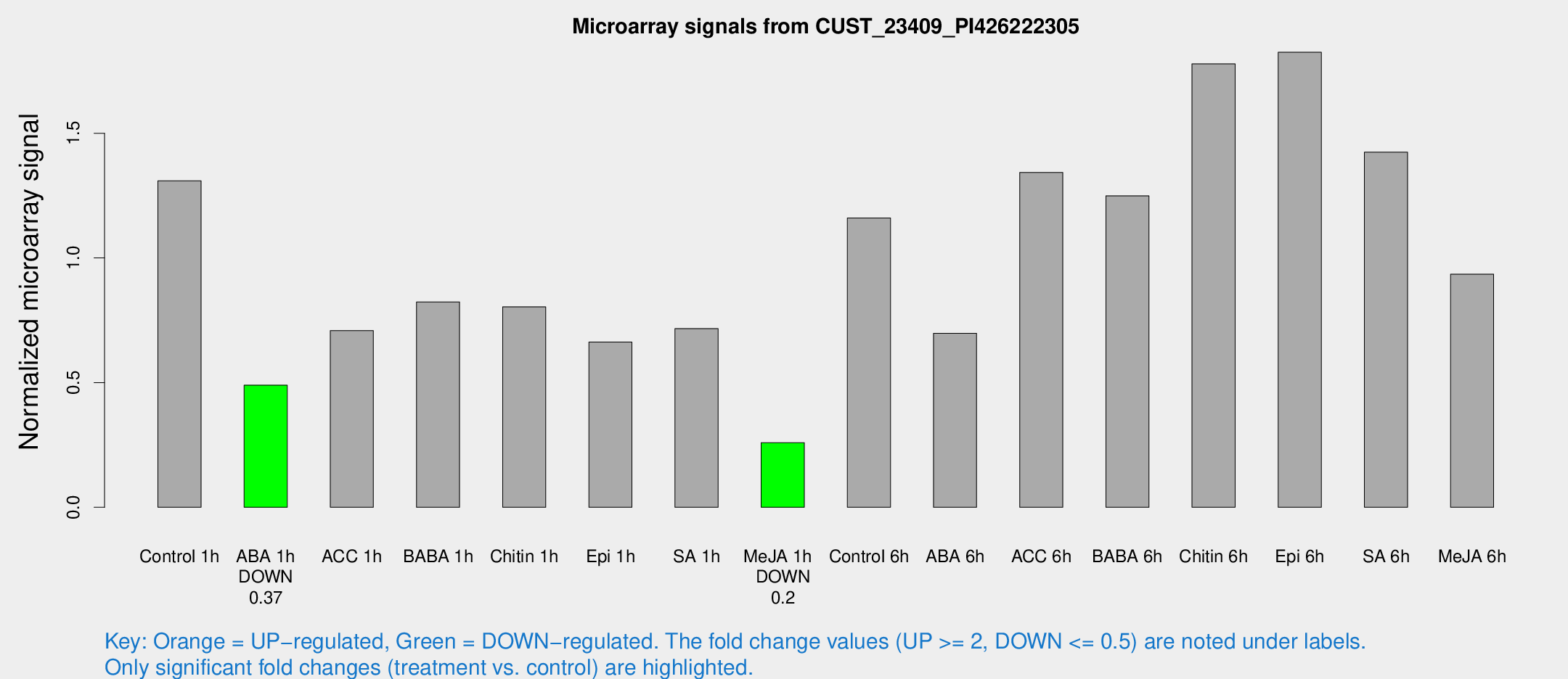
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 62.9766 | 10.7656 | 1.30973 | 0.132217 |
| ABA 1h | 20.6094 | 3.54253 | 0.489945 | 0.087569 |
| ACC 1h | 39.9239 | 13.3131 | 0.708999 | 0.279628 |
| BABA 1h | 38.6717 | 7.94117 | 0.823753 | 0.177267 |
| Chitin 1h | 35.6055 | 7.47606 | 0.804028 | 0.153574 |
| Epi 1h | 28.0569 | 5.50557 | 0.662645 | 0.142692 |
| SA 1h | 35.5002 | 5.76196 | 0.716863 | 0.177795 |
| Me-JA 1h | 9.90417 | 3.61201 | 0.259177 | 0.0948119 |
| Control 6h | 63.824 | 20.6343 | 1.16048 | 0.438229 |
| ABA 6h | 37.6267 | 11.0504 | 0.698012 | 0.251584 |
| ACC 6h | 75.3189 | 14.4792 | 1.34307 | 0.213438 |
| BABA 6h | 66.884 | 9.47841 | 1.24924 | 0.150342 |
| Chitin 6h | 88.7664 | 6.58285 | 1.77836 | 0.131995 |
| Epi 6h | 102.267 | 24.4939 | 1.82511 | 0.567639 |
| SA 6h | 66.4628 | 6.22648 | 1.42439 | 0.133939 |
| Me-JA 6h | 44.0566 | 4.71976 | 0.935158 | 0.0943173 |
Source Transcript PGSC0003DMT400065697 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G48870.1 | +3 | 5e-130 | 410 | 254/657 (39%) | Transducin/WD40 repeat-like superfamily protein | chr1:18072325-18074457 REVERSE LENGTH=593 |
