Probe CUST_22827_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22827_PI426222305 | JHI_St_60k_v1 | DMT400078243 | CTCAAGTGGATCGTAGATCCTTTGTAGCAATGCAAATTAAATCCTTCCTTGATTAATGAT |
All Microarray Probes Designed to Gene DMG400030450
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22576_PI426222305 | JHI_St_60k_v1 | DMT400078244 | CTCAAGTGGATCGTAGATCCTTTGTAGCAATGCAAATTAAATCCTTCCTTGATTAATGAT |
| CUST_22827_PI426222305 | JHI_St_60k_v1 | DMT400078243 | CTCAAGTGGATCGTAGATCCTTTGTAGCAATGCAAATTAAATCCTTCCTTGATTAATGAT |
Microarray Signals from CUST_22827_PI426222305
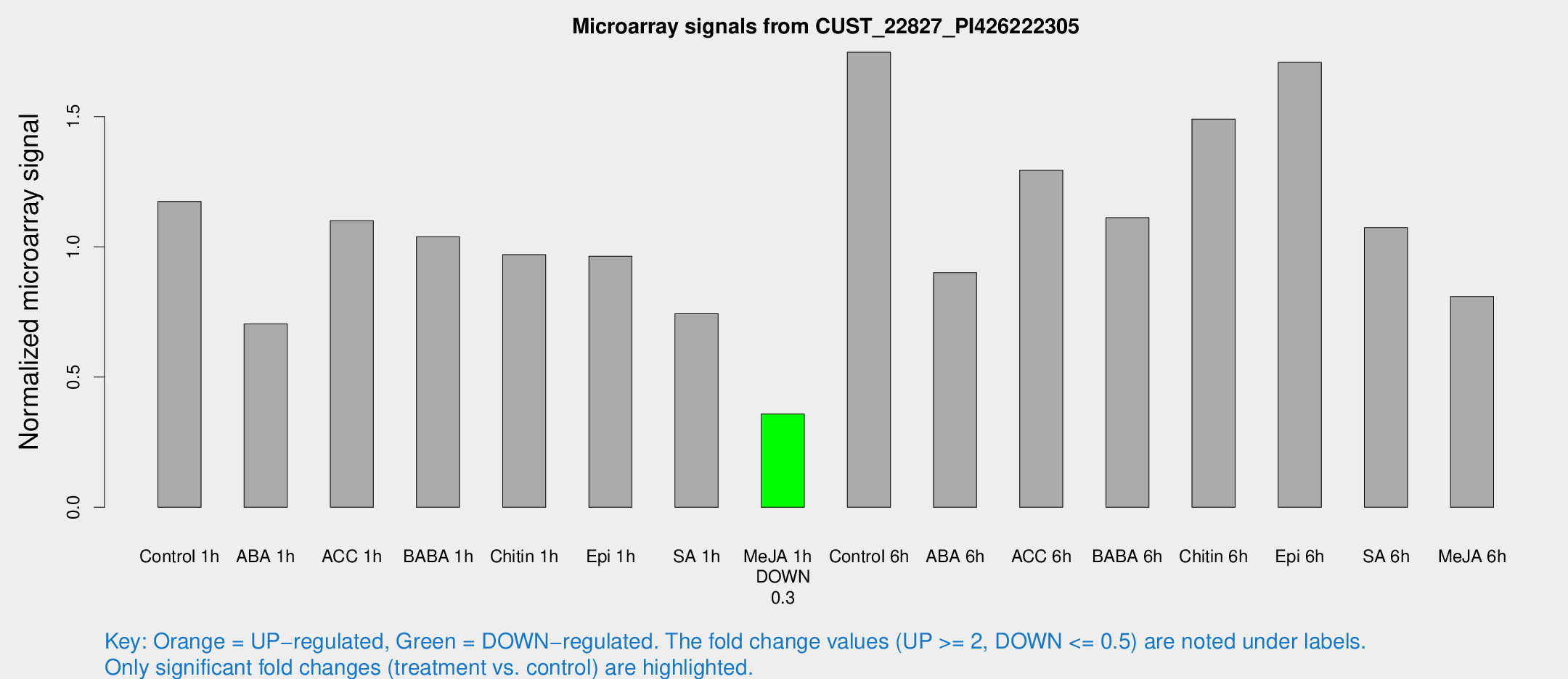
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 345.348 | 55.5764 | 1.17437 | 0.117408 |
| ABA 1h | 181.754 | 25.5791 | 0.704585 | 0.0576642 |
| ACC 1h | 347.819 | 95.0092 | 1.10015 | 0.230435 |
| BABA 1h | 294.797 | 51.1408 | 1.03836 | 0.121608 |
| Chitin 1h | 258.211 | 45.9537 | 0.970217 | 0.10355 |
| Epi 1h | 239.469 | 14.1856 | 0.963788 | 0.0569606 |
| SA 1h | 221.807 | 25.3642 | 0.743467 | 0.0555611 |
| Me-JA 1h | 83.8031 | 5.72841 | 0.357939 | 0.0244143 |
| Control 6h | 549.124 | 153.542 | 1.74739 | 0.397138 |
| ABA 6h | 276.284 | 23.812 | 0.901083 | 0.0626491 |
| ACC 6h | 436.236 | 72.6168 | 1.29474 | 0.0755344 |
| BABA 6h | 366.551 | 58.9931 | 1.11198 | 0.158765 |
| Chitin 6h | 454.862 | 26.5194 | 1.49037 | 0.0867708 |
| Epi 6h | 590.572 | 137.016 | 1.70816 | 0.64199 |
| SA 6h | 316.496 | 65.9547 | 1.07397 | 0.174701 |
| Me-JA 6h | 257.933 | 72.7438 | 0.809744 | 0.244238 |
Source Transcript PGSC0003DMT400078243 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G58520.1 | +1 | 0.0 | 766 | 390/602 (65%) | Protein kinase superfamily protein | chr5:23655312-23657943 FORWARD LENGTH=604 |
