Probe CUST_22496_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22496_PI426222305 | JHI_St_60k_v1 | DMT400039522 | ATTTCTGACTTGCAAGTGTATAAAATGCAAACAGTTCTCCGGAAACTATTCAAATGCTAG |
All Microarray Probes Designed to Gene DMG400015281
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22489_PI426222305 | JHI_St_60k_v1 | DMT400039518 | ATTTCTGACTTGCAAGTGTATAAAATGCAAACAGTTCTCCGGAAACTATTCAAATGCTAG |
| CUST_22496_PI426222305 | JHI_St_60k_v1 | DMT400039522 | ATTTCTGACTTGCAAGTGTATAAAATGCAAACAGTTCTCCGGAAACTATTCAAATGCTAG |
Microarray Signals from CUST_22496_PI426222305
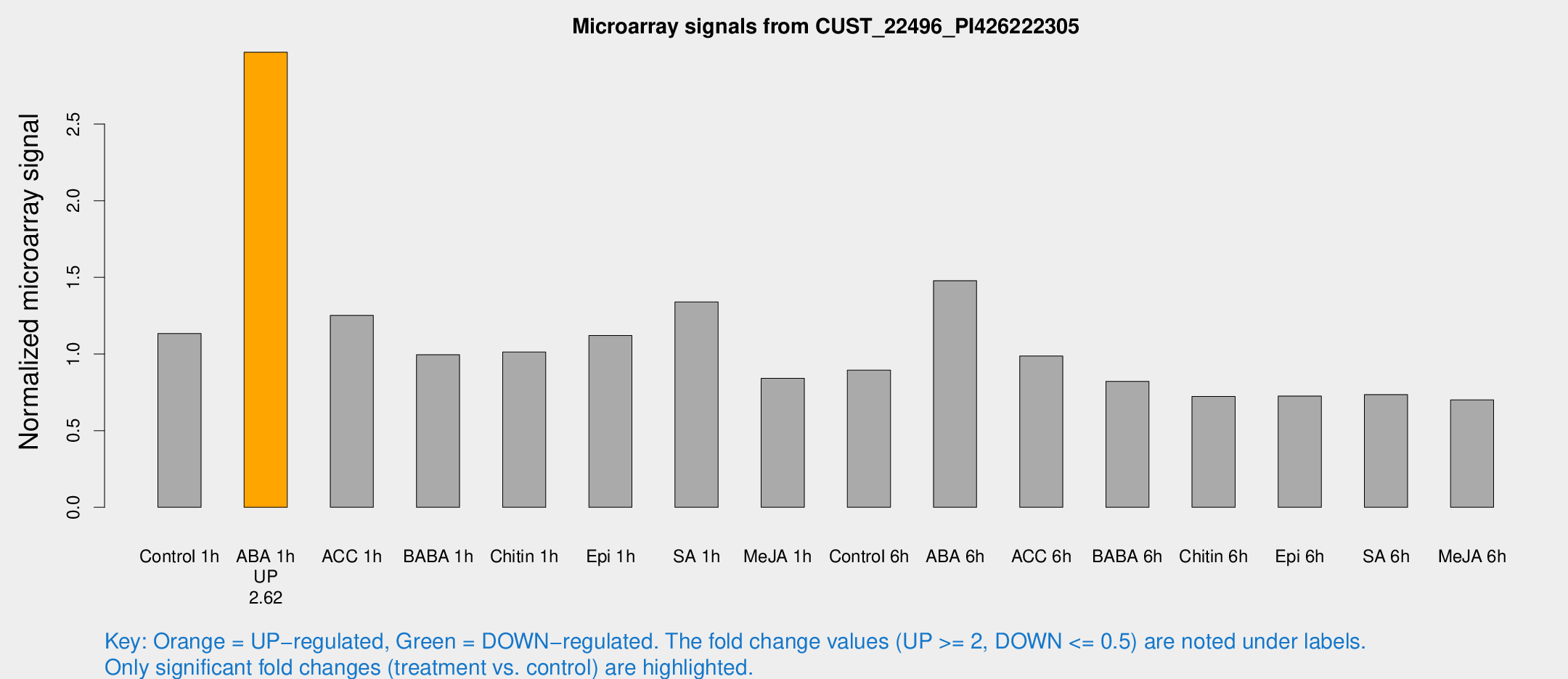
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 123.485 | 25.9329 | 1.13346 | 0.168122 |
| ABA 1h | 275.161 | 16.1792 | 2.96842 | 0.278535 |
| ACC 1h | 137.978 | 20.4547 | 1.25234 | 0.10517 |
| BABA 1h | 104.014 | 18.3729 | 0.995123 | 0.0921573 |
| Chitin 1h | 98.219 | 17.2188 | 1.01244 | 0.16123 |
| Epi 1h | 101.55 | 6.59384 | 1.12118 | 0.0727686 |
| SA 1h | 144.436 | 8.89708 | 1.33976 | 0.0823958 |
| Me-JA 1h | 72.0858 | 5.22103 | 0.84103 | 0.0908141 |
| Control 6h | 101.814 | 26.4991 | 0.894169 | 0.188894 |
| ABA 6h | 171.126 | 33.8169 | 1.47801 | 0.218903 |
| ACC 6h | 119.7 | 12.1161 | 0.987002 | 0.0648907 |
| BABA 6h | 96.6084 | 6.66523 | 0.821603 | 0.0566399 |
| Chitin 6h | 80.9286 | 5.94188 | 0.722666 | 0.0524681 |
| Epi 6h | 91.8515 | 25.3779 | 0.725595 | 0.213068 |
| SA 6h | 76.9316 | 8.2778 | 0.735061 | 0.103887 |
| Me-JA 6h | 76.2854 | 14.9617 | 0.700761 | 0.0935656 |
Source Transcript PGSC0003DMT400039522 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G73390.1 | +3 | 0.0 | 659 | 338/413 (82%) | Endosomal targeting BRO1-like domain-containing protein | chr1:27591079-27594148 REVERSE LENGTH=419 |
