Probe CUST_22352_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22352_PI426222305 | JHI_St_60k_v1 | DMT400039410 | ACTACAGATCTATAACAGCAAAGAACTTCCTGACATCTGTTAGAAGGTATCAAGGGGAGC |
All Microarray Probes Designed to Gene DMG400015241
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22352_PI426222305 | JHI_St_60k_v1 | DMT400039410 | ACTACAGATCTATAACAGCAAAGAACTTCCTGACATCTGTTAGAAGGTATCAAGGGGAGC |
| CUST_22375_PI426222305 | JHI_St_60k_v1 | DMT400039411 | TAAACTACAAGTACCATGAGTTAAGAAAATCATTCTCGTTCCCTAACTGGTTGTCATCAC |
Microarray Signals from CUST_22352_PI426222305
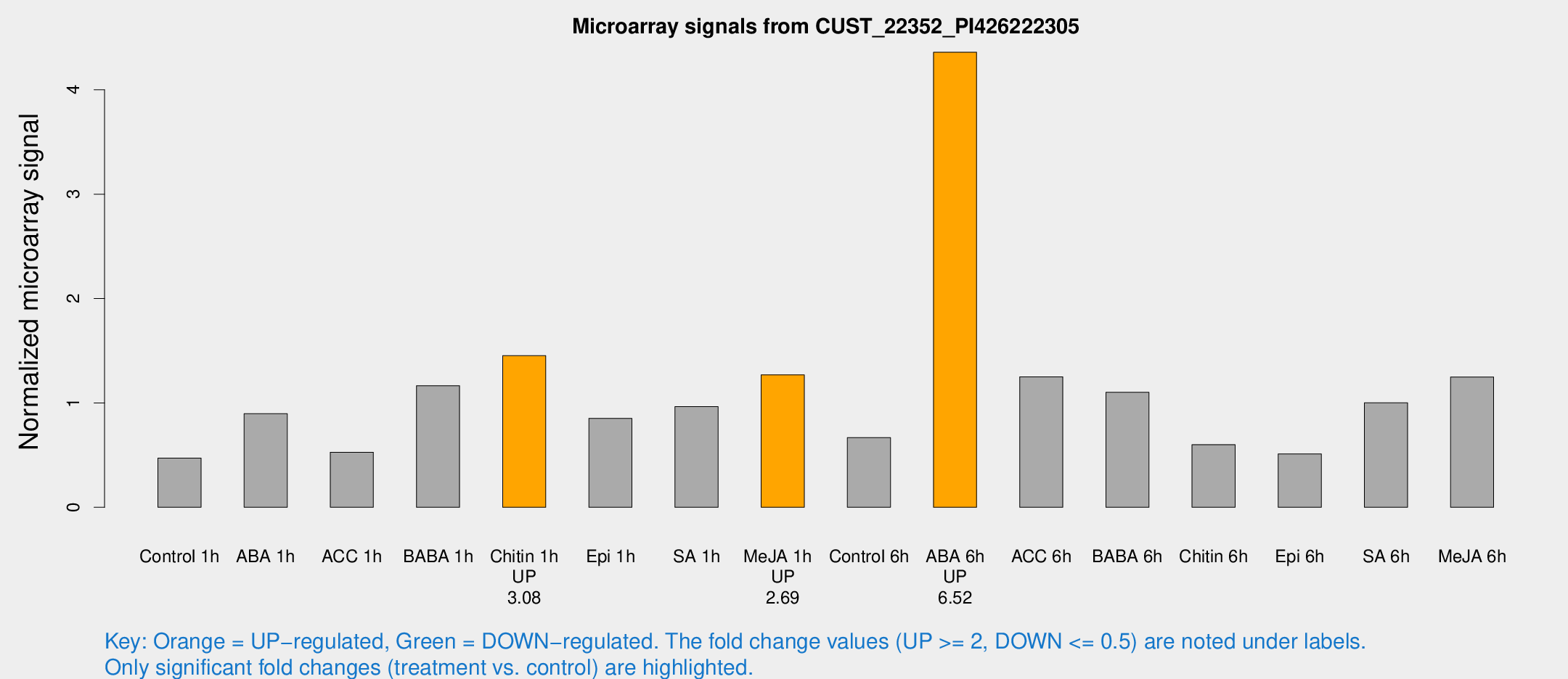
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 219.824 | 15.4097 | 0.471985 | 0.0281745 |
| ABA 1h | 428.805 | 140.749 | 0.896868 | 0.35981 |
| ACC 1h | 269.55 | 64.8525 | 0.527737 | 0.111058 |
| BABA 1h | 596.671 | 184.246 | 1.16502 | 0.367124 |
| Chitin 1h | 650.772 | 183.7 | 1.4528 | 0.288061 |
| Epi 1h | 343.468 | 23.1357 | 0.852065 | 0.0546229 |
| SA 1h | 532.674 | 207.632 | 0.964074 | 0.334468 |
| Me-JA 1h | 513.933 | 125.148 | 1.2693 | 0.239004 |
| Control 6h | 369.22 | 138.858 | 0.6683 | 0.264285 |
| ABA 6h | 2279.08 | 503.643 | 4.36018 | 0.913411 |
| ACC 6h | 669.103 | 54.4411 | 1.25026 | 0.0734606 |
| BABA 6h | 590.983 | 109.205 | 1.10145 | 0.160878 |
| Chitin 6h | 296.54 | 17.5839 | 0.600362 | 0.0355672 |
| Epi 6h | 305.27 | 92.4578 | 0.511828 | 0.182569 |
| SA 6h | 504.353 | 138.574 | 1.00141 | 0.213857 |
| Me-JA 6h | 581.645 | 52.8526 | 1.24892 | 0.0725467 |
Source Transcript PGSC0003DMT400039410 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G48530.1 | +3 | 5e-165 | 479 | 276/417 (66%) | SNF1-related protein kinase regulatory subunit gamma 1 | chr3:17987559-17989592 FORWARD LENGTH=424 |
