Probe CUST_22289_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22289_PI426222305 | JHI_St_60k_v1 | DMT400047621 | ATTCTGACTCTACTGCTACCGCGCAACTTATGGAAACGAATAAACTTTGTTTTGTTTTTT |
All Microarray Probes Designed to Gene DMG400018509
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22211_PI426222305 | JHI_St_60k_v1 | DMT400047622 | GGGGTGTAGAATAGGTTGTATATTCCAACTCAAGTAGAAGTCAAGGAATTTTAATATGTG |
| CUST_22289_PI426222305 | JHI_St_60k_v1 | DMT400047621 | ATTCTGACTCTACTGCTACCGCGCAACTTATGGAAACGAATAAACTTTGTTTTGTTTTTT |
Microarray Signals from CUST_22289_PI426222305
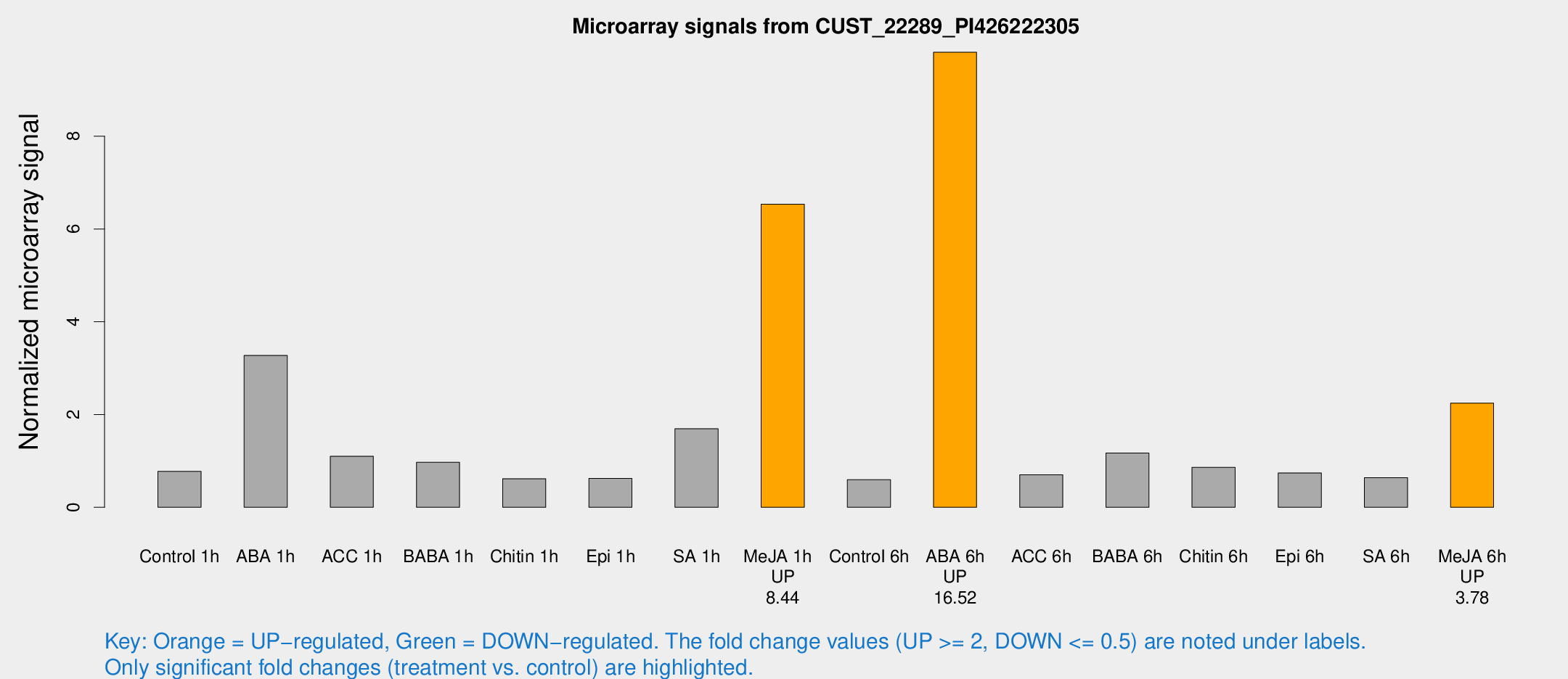
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.37541 | 3.74321 | 0.774759 | 0.36687 |
| ABA 1h | 45.1616 | 19.2221 | 3.27494 | 2.67332 |
| ACC 1h | 13.0536 | 4.7884 | 1.10103 | 0.432713 |
| BABA 1h | 11.3018 | 3.97712 | 0.970142 | 0.403124 |
| Chitin 1h | 6.28632 | 3.67702 | 0.616215 | 0.357581 |
| Epi 1h | 6.12177 | 3.57244 | 0.624802 | 0.362684 |
| SA 1h | 20.042 | 3.88484 | 1.69351 | 0.351658 |
| Me-JA 1h | 61.9005 | 10.2528 | 6.53582 | 0.627143 |
| Control 6h | 6.71497 | 3.90289 | 0.593919 | 0.344432 |
| ABA 6h | 123.632 | 26.106 | 9.81014 | 2.0465 |
| ACC 6h | 9.23962 | 4.47073 | 0.701973 | 0.345213 |
| BABA 6h | 14.8803 | 4.40068 | 1.17109 | 0.347442 |
| Chitin 6h | 10.897 | 4.35298 | 0.862767 | 0.381964 |
| Epi 6h | 9.42924 | 4.65791 | 0.741742 | 0.356272 |
| SA 6h | 7.13112 | 4.13115 | 0.640275 | 0.370807 |
| Me-JA 6h | 26.7528 | 6.933 | 2.24586 | 0.566581 |
Source Transcript PGSC0003DMT400047621 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G46680.1 | +1 | 2e-35 | 132 | 66/103 (64%) | homeobox 7 | chr2:19165777-19166773 REVERSE LENGTH=258 |
