Probe CUST_21162_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_21162_PI426222305 | JHI_St_60k_v1 | DMT400020227 | CCCTTTACTGAAAGGGCACACATTTAAAATTCAGCTTCTGGTAGCATAAACTTTAGATTA |
All Microarray Probes Designed to Gene DMG400007819
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_21083_PI426222305 | JHI_St_60k_v1 | DMT400020228 | CCCTTTACTGAAAGGGCACACATTTAAAATTCAGCTTCTGGTAGCATAAACTTTAGATTA |
| CUST_21162_PI426222305 | JHI_St_60k_v1 | DMT400020227 | CCCTTTACTGAAAGGGCACACATTTAAAATTCAGCTTCTGGTAGCATAAACTTTAGATTA |
Microarray Signals from CUST_21162_PI426222305
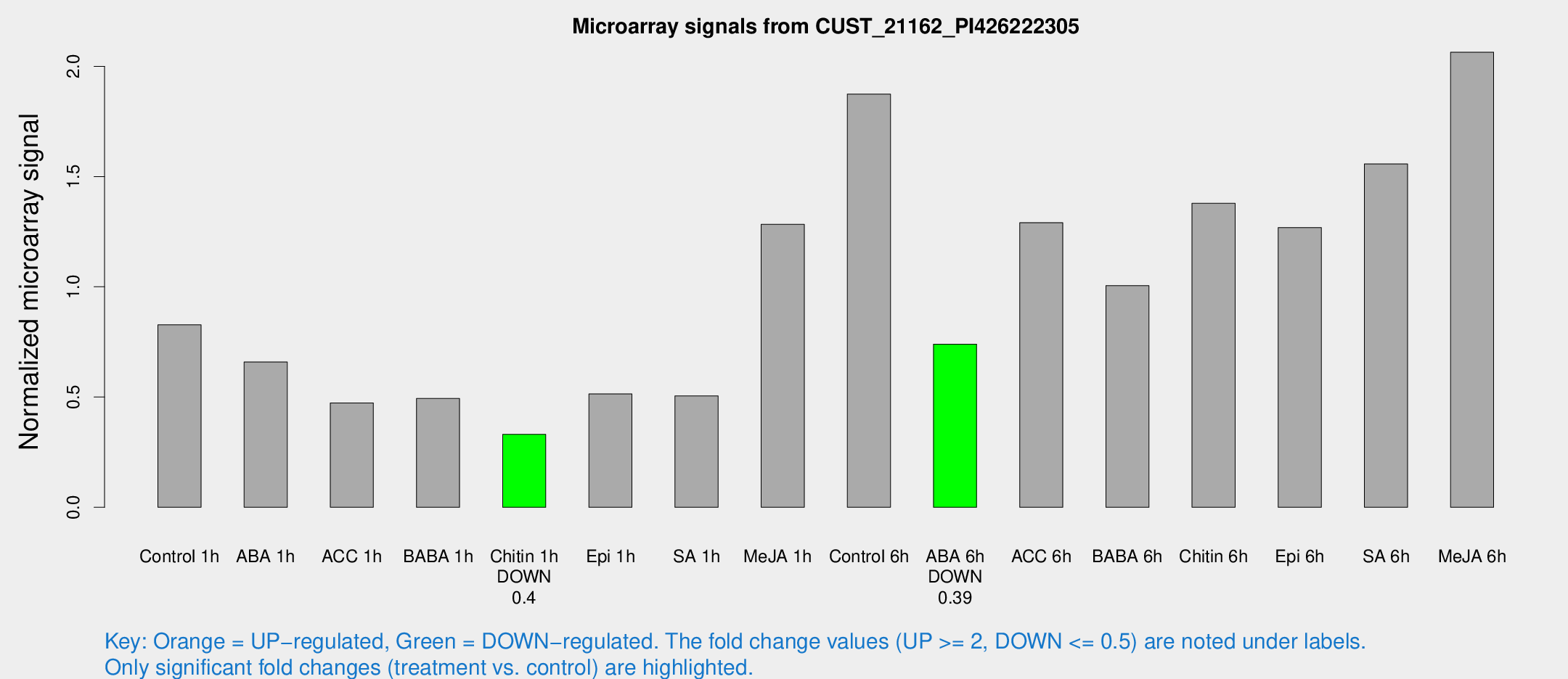
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 838.761 | 195.021 | 0.827771 | 0.134409 |
| ABA 1h | 567.011 | 53.2115 | 0.658667 | 0.061235 |
| ACC 1h | 611.845 | 290.555 | 0.472991 | 0.269098 |
| BABA 1h | 477.525 | 88.1022 | 0.49358 | 0.0512823 |
| Chitin 1h | 287.597 | 16.8912 | 0.330845 | 0.0193876 |
| Epi 1h | 431.457 | 30.5758 | 0.514004 | 0.0304499 |
| SA 1h | 555.029 | 186.427 | 0.505071 | 0.165822 |
| Me-JA 1h | 1027.14 | 132.693 | 1.28368 | 0.074211 |
| Control 6h | 1852.08 | 267.79 | 1.87423 | 0.115715 |
| ABA 6h | 770.993 | 93.1998 | 0.740074 | 0.0561486 |
| ACC 6h | 1432.82 | 83.048 | 1.29083 | 0.118182 |
| BABA 6h | 1098.46 | 122.2 | 1.00534 | 0.11361 |
| Chitin 6h | 1419.46 | 82.2518 | 1.37844 | 0.0796509 |
| Epi 6h | 1398.47 | 165.609 | 1.26884 | 0.168471 |
| SA 6h | 1489.49 | 86.266 | 1.55696 | 0.109453 |
| Me-JA 6h | 2003.11 | 202.46 | 2.06386 | 0.214354 |
Source Transcript PGSC0003DMT400020227 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G25620.1 | +1 | 2e-114 | 348 | 187/356 (53%) | DNA-binding protein phosphatase 1 | chr2:10903154-10904978 REVERSE LENGTH=392 |
