Probe CUST_20924_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20924_PI426222305 | JHI_St_60k_v1 | DMT400011762 | TGTCCCCTTCTAGATAAGTCTTTACTATAACCACTGAAAACACAAAGCAAGAGATGATTA |
All Microarray Probes Designed to Gene DMG400004617
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20770_PI426222305 | JHI_St_60k_v1 | DMT400011761 | CTCCTCAAGCTTGGAAAGATCCTTTGAAGAATTGTGCCTTTTCATATAAGGTAATGTTAT |
| CUST_20924_PI426222305 | JHI_St_60k_v1 | DMT400011762 | TGTCCCCTTCTAGATAAGTCTTTACTATAACCACTGAAAACACAAAGCAAGAGATGATTA |
Microarray Signals from CUST_20924_PI426222305
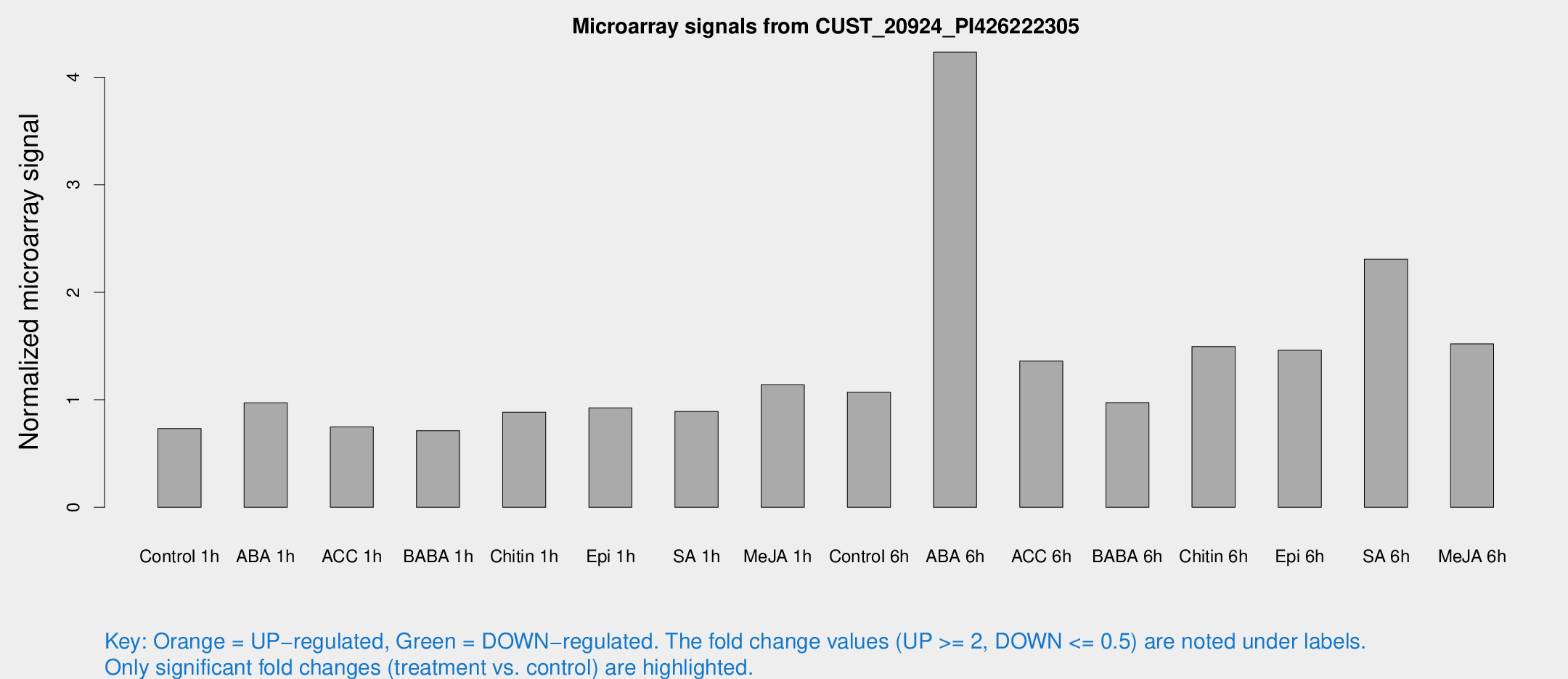
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6.84616 | 3.41218 | 0.732284 | 0.378856 |
| ABA 1h | 8.44413 | 3.27966 | 0.972066 | 0.433981 |
| ACC 1h | 7.13937 | 4.21853 | 0.747535 | 0.432988 |
| BABA 1h | 6.29239 | 3.66473 | 0.713005 | 0.413754 |
| Chitin 1h | 7.42737 | 3.41094 | 0.883886 | 0.415966 |
| Epi 1h | 7.65445 | 3.31498 | 0.924242 | 0.432278 |
| SA 1h | 9.99863 | 4.43395 | 0.891888 | 0.416466 |
| Me-JA 1h | 9.50911 | 3.5922 | 1.13876 | 0.536077 |
| Control 6h | 11.1903 | 4.2858 | 1.07095 | 0.448258 |
| ABA 6h | 45.7342 | 15.537 | 4.23292 | 1.26528 |
| ACC 6h | 15.1826 | 4.26496 | 1.36094 | 0.416796 |
| BABA 6h | 10.3688 | 3.94145 | 0.973863 | 0.404302 |
| Chitin 6h | 15.1871 | 3.9881 | 1.49472 | 0.431585 |
| Epi 6h | 16.7808 | 5.39234 | 1.46113 | 0.49823 |
| SA 6h | 21.5815 | 4.15566 | 2.30811 | 0.455011 |
| Me-JA 6h | 16.0212 | 5.1139 | 1.51967 | 0.581925 |
Source Transcript PGSC0003DMT400011762 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G47960.1 | +2 | 4e-27 | 107 | 61/145 (42%) | cell wall / vacuolar inhibitor of fructosidase 1 | chr1:17681954-17683516 REVERSE LENGTH=205 |
