Probe CUST_20748_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20748_PI426222305 | JHI_St_60k_v1 | DMT400011901 | GCAGCTATCTTCTAAAGGATCAACTCATGATGAAATTGACTTTGAGTTCTTAGGAAATGT |
All Microarray Probes Designed to Gene DMG400004670
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20748_PI426222305 | JHI_St_60k_v1 | DMT400011901 | GCAGCTATCTTCTAAAGGATCAACTCATGATGAAATTGACTTTGAGTTCTTAGGAAATGT |
| CUST_20742_PI426222305 | JHI_St_60k_v1 | DMT400011902 | TCTATGGACAAACTCTCTGGCTCTGGTTTTCAATCCAAAAAAGATTATCTTTTTGGGAGA |
Microarray Signals from CUST_20748_PI426222305
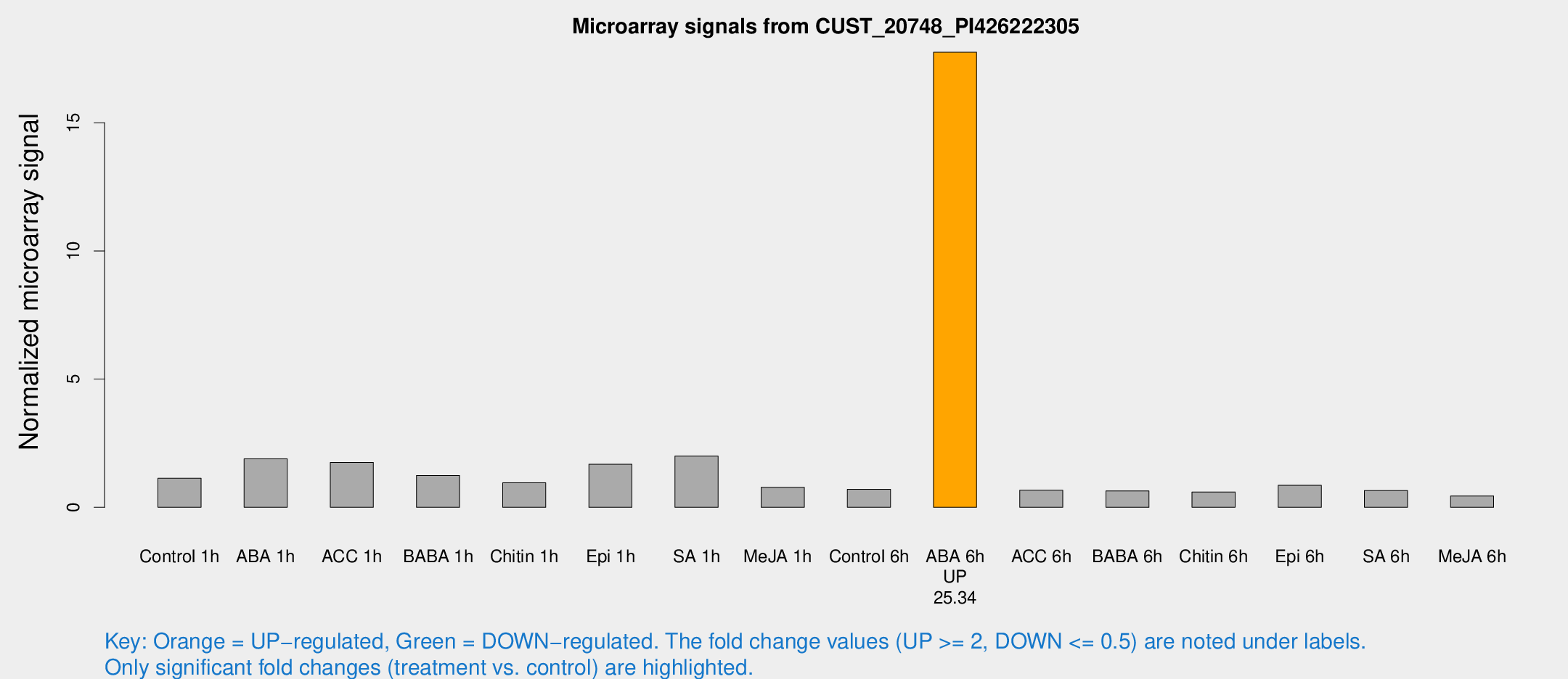
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 22.6399 | 10.4474 | 1.13309 | 0.464622 |
| ABA 1h | 28.7718 | 5.87335 | 1.88806 | 0.608349 |
| ACC 1h | 46.493 | 23.9244 | 1.74679 | 1.85776 |
| BABA 1h | 19.9694 | 3.4374 | 1.24222 | 0.30772 |
| Chitin 1h | 14.7528 | 3.22115 | 0.956148 | 0.226837 |
| Epi 1h | 23.9811 | 3.3464 | 1.68038 | 0.236809 |
| SA 1h | 36.4738 | 10.5432 | 1.99993 | 0.453957 |
| Me-JA 1h | 11.8946 | 3.9343 | 0.77947 | 0.308842 |
| Control 6h | 13.0027 | 4.12826 | 0.700496 | 0.246021 |
| ABA 6h | 328.426 | 77.8832 | 17.7488 | 4.23245 |
| ACC 6h | 12.5825 | 3.98391 | 0.665531 | 0.215371 |
| BABA 6h | 15.7892 | 8.69973 | 0.639888 | 0.386351 |
| Chitin 6h | 11.4495 | 3.62568 | 0.599313 | 0.231784 |
| Epi 6h | 16.2844 | 3.83869 | 0.858911 | 0.206992 |
| SA 6h | 12.2424 | 4.59496 | 0.653977 | 0.290628 |
| Me-JA 6h | 7.35941 | 3.23468 | 0.441653 | 0.204964 |
Source Transcript PGSC0003DMT400011901 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G14130.1 | +3 | 1e-105 | 313 | 148/195 (76%) | xyloglucan endotransglucosylase/hydrolase 15 | chr4:8137161-8138196 REVERSE LENGTH=289 |
