Probe CUST_19588_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19588_PI426222305 | JHI_St_60k_v1 | DMT400023252 | CTGGTTTTCTCAAAAGGGAACTTCCTGGATGAATGAAATCCAAATAGATTTACAATTTGG |
All Microarray Probes Designed to Gene DMG400009005
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19609_PI426222305 | JHI_St_60k_v1 | DMT400023251 | AGATTGTATGCTCAGATGTCTTATTTGGGAAGTTTGGCTTATGGAATACCCCAAATTAAG |
| CUST_19588_PI426222305 | JHI_St_60k_v1 | DMT400023252 | CTGGTTTTCTCAAAAGGGAACTTCCTGGATGAATGAAATCCAAATAGATTTACAATTTGG |
Microarray Signals from CUST_19588_PI426222305
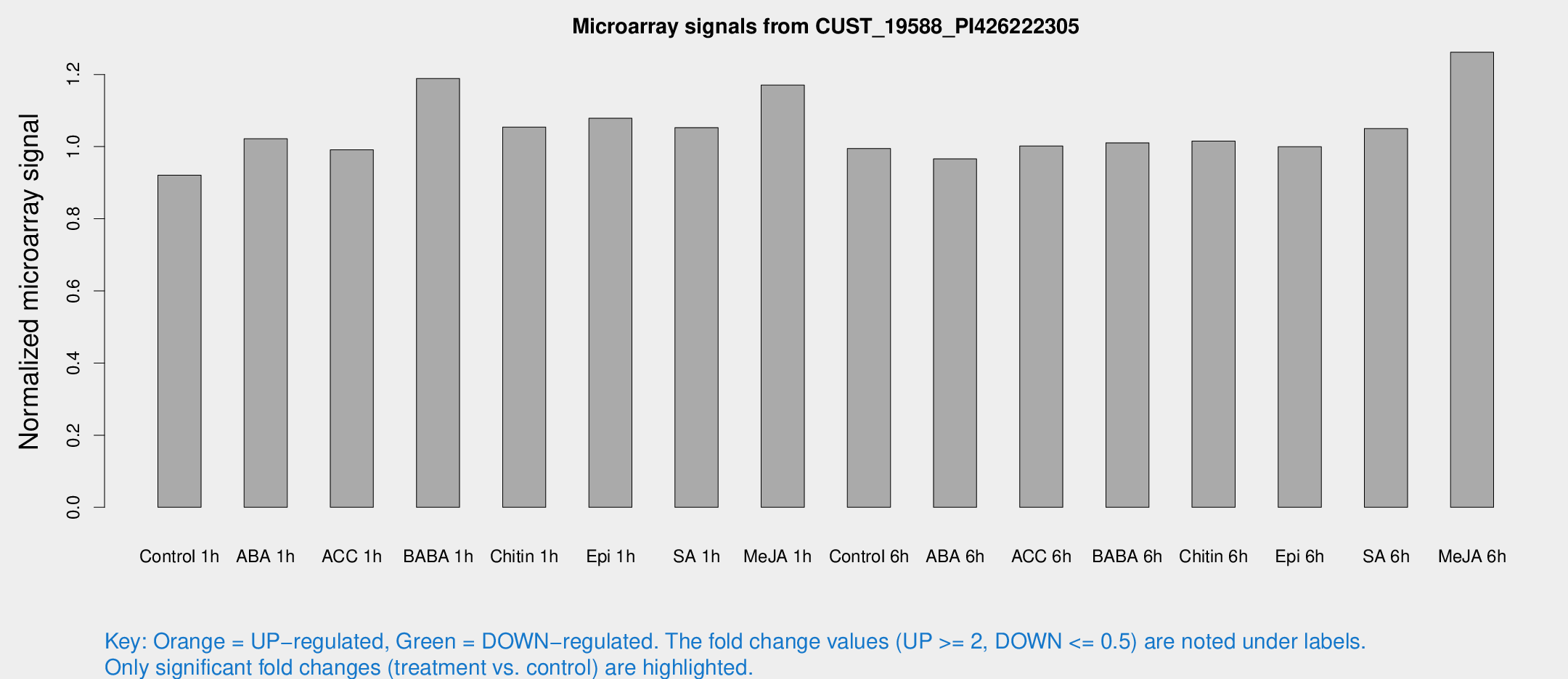
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.88091 | 3.40639 | 0.9206 | 0.533075 |
| ABA 1h | 5.78266 | 3.35461 | 1.0219 | 0.591847 |
| ACC 1h | 6.51429 | 3.78118 | 0.990906 | 0.573808 |
| BABA 1h | 7.50345 | 3.70264 | 1.18873 | 0.613282 |
| Chitin 1h | 6.07166 | 3.52594 | 1.0542 | 0.610473 |
| Epi 1h | 5.97097 | 3.46737 | 1.07838 | 0.625167 |
| SA 1h | 7.1205 | 3.46231 | 1.05247 | 0.539338 |
| Me-JA 1h | 6.11559 | 3.55127 | 1.1706 | 0.678266 |
| Control 6h | 6.36959 | 3.63854 | 0.99479 | 0.567653 |
| ABA 6h | 6.56497 | 3.80629 | 0.9659 | 0.559464 |
| ACC 6h | 7.53316 | 4.48409 | 1.00169 | 0.580047 |
| BABA 6h | 7.27078 | 4.11222 | 1.01043 | 0.571853 |
| Chitin 6h | 6.90076 | 3.99738 | 1.01503 | 0.587942 |
| Epi 6h | 7.25765 | 4.23951 | 0.999553 | 0.578967 |
| SA 6h | 6.63622 | 3.8444 | 1.05013 | 0.608119 |
| Me-JA 6h | 8.56624 | 3.70077 | 1.26159 | 0.615895 |
Source Transcript PGSC0003DMT400023252 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G02660.1 | +3 | 2e-64 | 216 | 118/209 (56%) | alpha/beta-Hydrolases superfamily protein | chr1:572187-574746 REVERSE LENGTH=713 |
